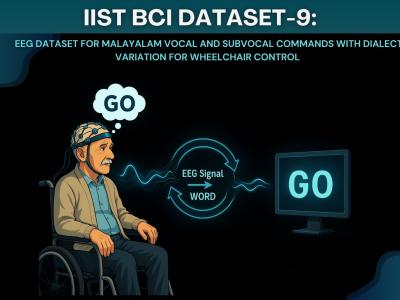
We developed IIST BCI Dataset-9, a novel EEG-based Brain-Computer Interface (BCI)
dataset to improve wheelchair control systems using Malayalam dialect variations. BCI
systems help people with motor disabilities by allowing them to control devices using brain
signals. The limited number of BCI datasets in Indian languages makes it harder for native
speakers to use these systems. To address this, we created a dataset with 15 Malayalam
words related to basic wheelchair commands like Forward, Backward, Go, Stop, Reverse,
- Categories: