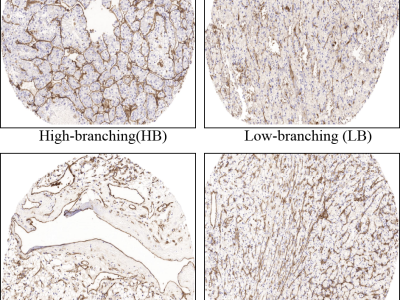
This dataset comprises 325 histopathological images of clear cell renal cell carcinoma (ccRCC) tissue sections, designed to characterize vascular morphology based on CD31 immunohistochemical staining. Each image was scanned at 10× magnification and annotated with global proportions for three distinct vascular patterns: high-branching (HB), low-branching (LB), and sinusoidal (SN). The provided annotations include the relative distribution of each vascular class per image.
- Categories: