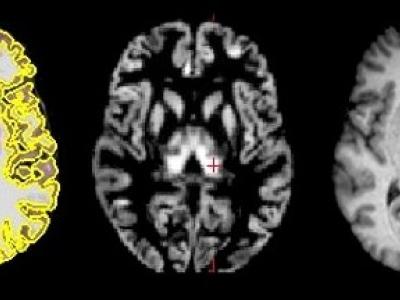
Design of EEG-TMS experiment. The figure shows the timeline of the experimental session, the illustration of the typical sequence of the visual cues and the structure of one trial, and the illustration of the time intervals of interest within a trial: Pre is the baseline pre-que interval [-4.5 -0.5] s; Post is the post-cue interval [0 0.5] s; Img is the interval [1 3] s of motor imagery execution; here, t=0 corresponds to the moment of the appearance of the visual cue to start the movement.
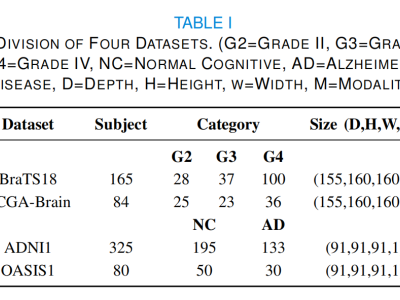
- Categories: