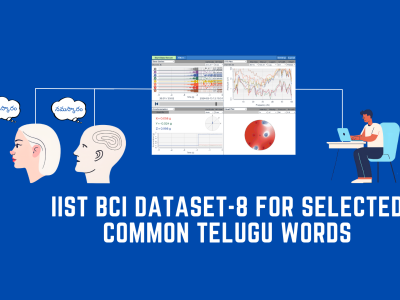
This dataset consists of carefully curated audio recordings that capture the distinct sounds produced by multiple individuals walking in various environments. Designed to support research in sound recognition, activity analysis, and the study of human behaviour, it provides a rich resource for understanding how group dynamics influence acoustic patterns. Each recording is accompanied by detailed metadata, including the number of participants, environmental context, and surface characteristics.
- Categories: