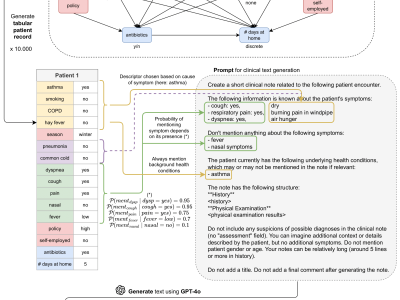
We present the SynSUM benchmark, a synthetic dataset linking unstructured clinical notes to structured background variables. The dataset consists of 10,000 artificial patient records containing tabular variables (like symptoms, diagnoses and underlying conditions) and associated clinical notes describing the fictional patient encounter in the domain of respiratory diseases.
- Categories:




