Machine Learning
Anomaly detection in Phasor Measurement Unit (PMU) data requires high-quality, realistic labeled datasets for algorithm training and validation. Obtaining real field labelled data is challenging due to privacy, security concerns, and the rarity of certain anomalies, making a robust testbed indispensable. This paper presents the development and implementation of a Hardware-in-the-Loop (HIL) Synchrophasor Testbed designed for realistic data generation for testing and validating PMU anomaly detection algorithms.
- Categories:
 1005 Views
1005 Views
Advancements in Medical Vision-Language Pre-training (Medical-VLP) progress rapidly by learning representations from paired radiology reports. Nevertheless, there still remain two issues that restrict the development of Medical-VLP: the scarcity of parallel image-report pair and monotony of pre-training tasks. Thus, we propose Multi-Grained Cross-Domain Report Searching (CDRS) strategy, and Multi-Task Driven Language-Image Pre-Training (MLIP) framework.
- Categories:
 52 Views
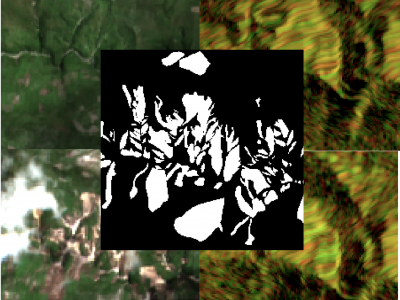
52 ViewsThe detection of the collapse of landslides trigerred by intense natural hazards, such as earthquakes and rainfall, allows rapid response to hazards which turned into disasters. The use of remote sensing imagery is mostly considered to cover wide areas and assess even more rapidly the threats. Yet, since optical images are sensitive to cloud coverage, their use is limited in case of emergency response. The proposed dataset is thus multimodal and targets the early detection of landslides following the disastrous earthquake which occurred in Haiti in 2021.
- Categories:
 691 Views
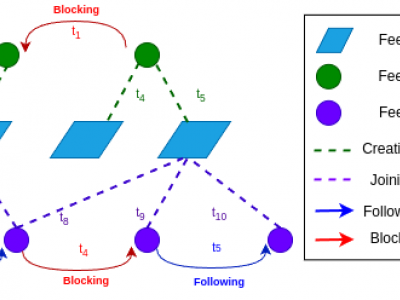
691 ViewsDecentralized social media platforms like Bluesky Social (Bluesky) have made it possible to publicly disclose some user behaviors with millisecond-level precision. Embracing Bluesky's principles of open-source and open-data, we present the first collection of the temporal dynamics of user-driven social interactions. BlueTempNet integrates multiple types of networks into a single multi-network, including user-to-user interactions (following and blocking users) and user-to-community interactions (creating and joining communities).
- Categories:
 974 Views
974 Views
Machine learning (ML) in the medical domain faces challenges due to limited high-quality data. This study addresses the scarcity of echocardiography images (echoCG) by generating synthetic data using state-of-the-art generative models. We evaluated a cycle-consistent generative adversarial network (CycleGAN), contrastive unpaired translation (CUT) method, and latent diffusion model (Stable Diffusion 1.5).
- Categories:
 145 Views
145 ViewsA dataset related to a cable under anomalies conditions while a transmitter (AWG) sends a binary PAM signal to a receiver. The signals are acquired by an oscilloscope. The anomalies were manually forced on the cable under test: air-exposed, water-exposed conductors, and tapping. In the dataset, the signals are also available for normal cable.
- Categories:
 365 Views
365 ViewsWe are pleased to introduce the Qilin Watermelon Dataset, a unique collection of data aimed at investigating the relationship between a watermelon's appearance, tapping sound, and sweetness. This dataset is the result of our dedicated efforts to capture and record various aspects of Qilin watermelons, a special variety known for its exceptional taste and quality.
- Categories:
 1330 Views
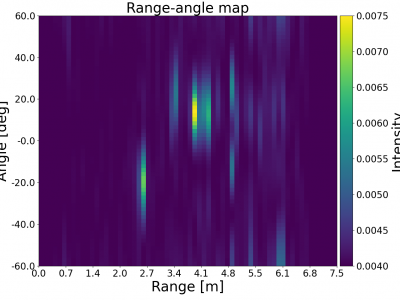
1330 ViewsIn our work, we propose an innovative system to accurately infer and track occluded target locations using mmWave beat frequency signals. Our approach combines a classic direction-finding method with advanced deep learning techniques, specifically a convolutional neural network (CNN), to enhance detection capabilities. The dataset includes raw beat frequency signal data from the TI IWR6843ISK rev B with TI mmWAVEICBOOST and the TI DCA1000EVM capture board. Corresponding ground truth data (target position) from the Realsense L515 RGB-D camera is also provided.
- Categories:
 333 Views
333 ViewsAs with most AI methods, a 3D deep neural network needs to be trained to properly interpret its input data. More specifically, training a network for monocular 3D point cloud reconstruction requires a large set of recognized high-quality data which can be challenging to obtain. Hence, this dataset contains the image of a known object alongside its corresponding 3D point cloud representation. To collect a large number of categorized 3D objects, we use the ShapeNetCore (https://shapenet.org) dataset.
- Categories:
 612 Views
612 Views
The dataset exemplifies land vehicle targets, tanks, and comprises 1000 time-frequency representation (TFR) images in jpg format with a resolution of 875x656 pixels. Each image is accompanied by labels containing 14 parameters for geometric parameter prediction.
- Categories:
 69 Views
69 Views