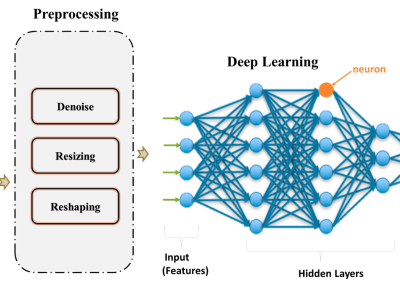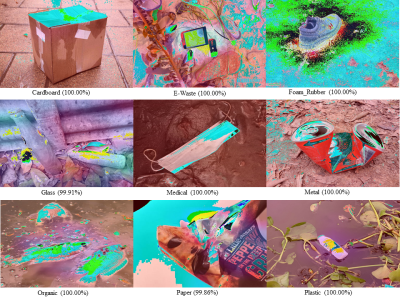
An automatic waste classification system embedded with higher accuracy and precision of convolution neural network (CNN) model can significantly the reduce manual labor involved in recycling. The ConvNeXt architecture has gained remarkable improvements in image recognition. A larger dataset, called TrashNeXt, comprising 23,625 images across nine categories has been introduced in this study by combining and thoroughly analyzing various pre-existing datasets.
- Categories: