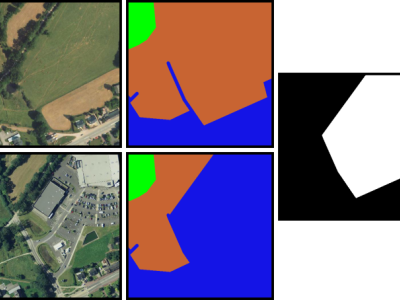
This dataset records the data required for the paper "Enhancing Stateful Processing in Programmable Data Planes: Model and Improved Architecture", comprising five .xlsx files, corresponding to Figures 3, 7, 19, 20, and 21 in the paper. Each data file includes notes that explain the meaning of the data, the headers of the rows and columns, and the units.
- Categories: