Medical Imaging

Elastography is a non-invasive technique to detect tissue anomalies via the local elastic modulus using shear waves. Commonly shear waves are produced via acoustic focusing or the use of mechanical external sources, shear waves may result also naturally from cavitation bubbles during medical intervention, for example from thermal ablation. Here, we measure the shear wave emitted from a well-controlled single laser-induced cavitation bubble oscillating near a rigid boundary. The bubbles are generated in a transparent tissue-mimicking hydrogel embedded with tracer particles.
- Categories:
 17 Views
17 Views
This simulation dataset contains five types of data: resolutions, vessels, vessel stenosis, tumors, and shape combinations. There are a total of 1000 original binary images. Besides, we set different gray values on images with multiple connected domains to simulate different concentration of magnetic nanoparticles. Next, the images are subjected to operations such as image inversion and image rotation. The final dataset contains 20,000 images. we applied the X-space method based on the X-space theory and we generated the simulated image of magnetic particle imaging.
- Categories:
 342 Views
342 Views
- Categories:
 417 Views
417 ViewsThis Dataset used a non-invasive blood group prediction approach using deep learning. Rapid and meticulous prediction of blood type is a major step during medical emergency before supervising the red blood cell, platelet, and plasma transfusion. Any small mistake during transfer of blood can cause death. In conventional pathological assessment, the blood test is conducted using automated blood analyser; however, it results into time taking process.
- Categories:
 3366 Views
3366 Views
Tuberculosis (TB) remains a major global health problem with high incidence and mortality rates worldwide. In recent years, with the rapid development of computer-aided diagnosis (CAD) tools, CAD has played an increasingly important role in supporting tuberculosis diagnosis. However, the development of CAD for TB diagnosis relies heavily on well-annotated computerized tomography (CT) datasets. Unfortunately, the currently available annotations in TB CT datasets are still limited, which hinders the development of CAD tools for TB diagnosis to some extent.
- Categories:
 152 Views
152 Views
Tuberculosis (TB) remains a major global health problem with high incidence and mortality rates worldwide. In recent years, with the rapid development of computer-aided diagnosis (CAD) tools, CAD has played an increasingly important role in supporting tuberculosis diagnosis. However, the development of CAD for TB diagnosis relies heavily on well-annotated computerized tomography (CT) datasets. Unfortunately, the currently available annotations in TB CT datasets are still limited, which hinders the development of CAD tools for TB diagnosis to some extent.
- Categories:
 9 Views
9 Views
Accurate detection and segmentation of brain tumors is critical for medical diagnosis. We propose a novel framework Two-Stage Generative Model (TSGM) that combines Cycle Generative Adversarial Network (CycleGAN) and Variance Exploding stochastic differential equation using joint probability (VE-JP) to improve brain tumor segmentation. TSGM was trained on the BraTs2020 brain tumor dataset.
- Categories:
 73 Views
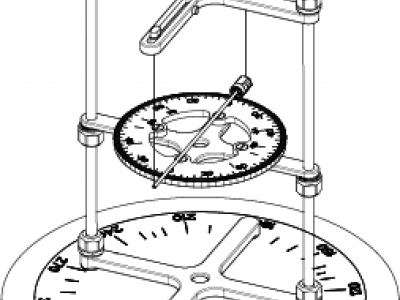
73 ViewsUse of medical devices in the magnetic resonance environment is regulated by standards that include the ASTM-F2213 magnetically induced torque. This standard prescribes five tests. However, none can be directly applied to measure very low torques of slender lightweight devices such as needles. Methods: We present a variant of an ASTM torsional spring method that makes a “spring” of 2 strings that suspend the needle by its ends. The magnetically induced torque on the needle causes it to rotate. The strings tilt and lift the needle.
- Categories:
 284 Views
284 Views
We introduce a high-performance computer vision based Intraveneous (IV) infusion speed measurement system as a camera application on an iPhone or Android phone. Our system uses You Only Look Once version 5 (YOLOv5) as it was designed for real-time object detection, making it substantially faster than two-stage algorithms such as R-CNN. In addition, YOLOv5 offers greater precision than its predecessors, making it more competitive with other object detection methods.
- Categories:
 65 Views
65 Views— Medical image segmentation is a crucial aspect of medical image processing, and has been widely used in the detection and clinical diagnosis for brain, lung, liver, heart and other diseases. In this paper, we propose a novel multimodal mutual attention network, called MMAUNet, for medical image segmentation. MMA-UNet is divided into two parts. The first part obtains more highdimensional features by skip connection and improved network structure.
- Categories:
 297 Views
297 Views