- Categories:

The accurate identification of miRNA-disease associations plays a crucial role in biomedical research and clinical applications. However, most research focuses on the existence of the association, without conducting further exploration. In this study, we propose a novel statistical meta-path contrastive learning-based approach (SMCLMDA), which aims to accurately identify the multidimensional relationships(up/down-regulation and causal/non-causal) between miRNAs and diseases.
- Categories:

Most promoters are derived from an arbitrary truncation of sequences upstream from the transcription start site of a gene, which is typically around 1,000 base pairs. Since the truncation is arbitrary, regulatory elements might be missing for transcription. Unfortunately, there exists no reasonable rationale for selecting a truncation threshold. Therefore, providing a reasonable rationale for truncation is crucial for obtaining the expected expression profiles of genes.
- Categories:

This study explores the potential of electromyography (EMG) decoding to enhance motor function outcomes, focusing on developing an innovative EMG-based hand movement classification system. Leveraging advanced signal processing and machine learning techniques, our objectives are twofold: (1) optimize EMG decoding performance through time-domain windowing, feature selection, and classifier optimization, and (2) assess the system's effectiveness in classifying 15 distinct finger movements.
- Categories:

We curated and release a real-world medical clinical dataset, namely MedCD, in the context of building generative artificial intelligence (AI) applications in the clinical setting. The MedCD dataset is one of the accomplishments from our longitudinal applied AI research and deployment in a tertiary care hospital in China. First, the dataset is real and comprehensive, in that it was sourced from real-world electronic health records (EHRs), clinical notes, lab examination reports and more.
- Categories:
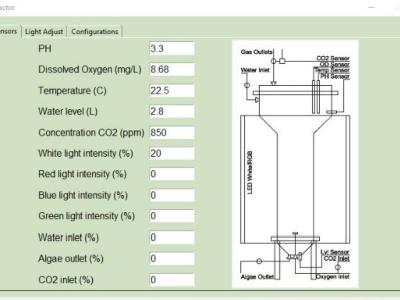
<p>Code used to read the sensors, and light control</p>
- Categories:

Antisense oligonucleotides (ASOs) have emerged as a new therapeutic modality for the treatment of both rare and common human diseases. A significant proportion of the patient population that may benefit from ASO therapy may also have common diseases, such as diabetes mellitus. The potential influence of prevalent diseases on the effectiveness of ASO drugs in silencing their target mRNAs remains largely unexplored. The current study utilized in vitro cell models to determine the impact on the FDA-approved ASO drug-mediated target reduction by cellular glucose levels.
- Categories:

The subject of the conducted project was research and development work aimed at developing a product innovation in the form of a robotic hospital ward. The task of such a ward was to minimize the participation of medical personnel in processes requiring contact with the patient, and thus the risk of infections and further spread of viruses.
- Categories:

This is a videoconference between a witness about murders who is a victim of many crimes and a law firm. This witness is called Colin Paul Gloster. This law firm is called Pais do Amaral Advogados.
\begin{quotation}``A significant part of the background of eh this process is that Hospital
Sobral Cid tortured me during 2013 eh but eh this process is actually
about consequences eh thru through a later process [. . .] To obscure eh
this torture of 2013, eh a new show trial was intitiated. Its process was
- Categories:

With the development of artificial intelligence, new possibilities have been opened up for the diagnosis and prevention of osteoporosis. We have successfully constructed an osteoporosis risk prediction model using deep learning algorithms, combined with demographic data and laboratory results. To further promote human health, we will publicly disclose the dataset we used, which includes 2,186 cases of males over 50 years old and postmenopausal females.
- Categories:
