Artificial Intelligence
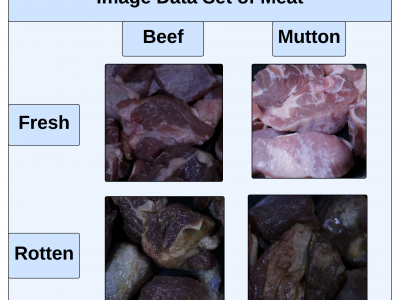
This paper presents an innovative Internet of Things (IoT) system that integrates gas sensors and a custom Convolutional Neural Network (CNN) to classify the freshness and species of beef and mutton in real time. The CNN, trained on 9,928 images, achieved 99% accuracy, outperforming models like ResNet-50, SVM, and KNN. The system uses three gas sensors (MQ135, MQ4, MQ136) to detect gases such as ammonia, methane, and hydrogen sulfide, which indicate meat spoilage.
- Categories:
 291 Views
291 Views
To provide a standardized approach for testing and benchmarking secure evaluation of transformer-based models, we developed the iDASH24 Homomorphic Encryption track dataset. This dataset is centered on protein sequence classification as the benchmark task. It includes a neural network model with a transformer architecture and a sample dataset, both used to build and evaluate secure evaluation strategies.
- Categories:
 130 Views
130 Views
The quality and safety of tea food production is of paramount importance. In traditional processing techniques, there is a risk of small foreign objects being mixed into Pu-erh sun-dried green tea, which directly affects the quality and safety of the food. To rapidly detect and accurately identify these small foreign objects in Pu-erh sun-dried green tea, this study proposes an improved YOLOv8 network model for foreign object detection. The method employs an MPDIoU optimized loss function to enhance target detection performance, thereby increasing the model's precision in targeting.
- Categories:
 68 Views
68 Views
The datasets are sourced from the Caltrans Performance Measurement System (PeMS) in California, which monitors and collects real-time traffic data from over 39,000 sensors deployed on major highways throughout the state. The PeMS system collects data every 30 seconds and aggregates it into 5-minute interval, with each sensor generating data for 288 time steps daily. Additionally, road network structure data is derived from the connectivity status and actual distances between sensors.
- Categories:
 178 Views
178 ViewsThe TiHAN-V2X Dataset was collected in Hyderabad, India, across various Vehicle-to-Everything (V2X) communication types, including Vehicle-to-Vehicle (V2V), Vehicle-to-Infrastructure (V2I), Infrastructure-to-Vehicle (I2V), and Vehicle-to-Cloud (V2C). The dataset offers comprehensive data for evaluating communication performance under different environmental and road conditions, including urban, rural, and highway scenarios.
- Categories:
 525 Views
525 Views
Abstract—Most reinforcement learning algorithms for robotic arm control in sparse reward environments are primarily optimized for end-effector displacement control mode.
- Categories:
 25 Views
25 Views
This dataset includes two 2D medical image segmentation benchmark.
1. OD/OC Segmentation in Fundus Image
This dataset conprises five sub-datasets: Drishti-GS, RIM-ONE-r3, ORIGA, REFUGE, and the validation set of REFUGE2. Each image is cropped around the optic disc area. The size of all images is 512×512. The manual pixel-wise annotation is stored as a PNG image with the same size as the corresponding fundus image with the following labels:
128: Optic Disc (Grey color)
0: Optic Cup (Black color)
255: Background (White color)
- Categories:
 50 Views
50 Views
"Recent advancements in deep learning and generative models have significantly enhanced text-to-image (T2I) synthesis, allowing for the creation of highly realistic images based on textual inputs. While this progress has expanded the creative and practical applications of AI, it also presents new challenges in distinguishing between authentic and AI-generated images. This challenge raises serious concerns in areas such as security, privacy, and digital forensics.
- Categories:
 190 Views
190 ViewsIn this research, a newly modified UNet (Fast-UNet) was implemented to segment winter wheat from time series Sentinel-2 images for the years 2021 and 2023. These images were converted to NDVI and utilized to identify wheat fields by tracking the wheat phenology from sowing to harvesting. The main satellite image that was used in this research was Sentinel-2. It is considered important, and free optical remote sensing satellite data is provided by the European Space Agency (ESA). Sentinel-2A and Sentinel-2B were launched in June 2015 and March 2017, respectively.
- Categories:
 197 Views
197 Views
The dataset is derived from the ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) domain, which plays a crucial role in drug discovery and development. This dataset comprises a comprehensive collection of chemical compounds with associated ADMET properties, providing a rich resource for evaluating the efficacy of machine learning models in predicting drug behavior. Specifically, the dataset includes diverse features representing molecular structures, physicochemical properties, and biological activity profiles, allowing for robust modeling of classification tasks.
- Categories:
 97 Views
97 Views