Artificial Intelligence

We present SMPL-IKS, an inverse kinematic solver to operate on the well-known Skinned Multi-Person Linear model (SMPL) to recover human mesh from 3D skeleton. The challenges of the task are threefold: (1) Shape Mismatching. (2) Error Accumulation. (3) Rotation Ambiguity. Instead of recovering human mesh from costly vertice up-sampling or iterative optimization as in previous methods, SMPL-IKS directly regresses the SMPL parameters (i.e., shape and pose parameters) in a clean and efficient way. Specifically, we propose to infer skeleton-to-mesh via two explicit mappings viz.
- Categories:
 61 Views
61 ViewsJamming devices pose a significant threat by disrupting signals from the global navigation satellite system (GNSS), compromising the robustness of accurate positioning. Detecting anomalies in frequency snapshots is crucial to counteract these interferences effectively. The ability to adapt to diverse, unseen interference characteristics is essential for ensuring the reliability of GNSS in real-world applications. We recorded a dataset with our own sensor station at a German highway with eight interference classes and three non-interference classes.
- Categories:
 150 Views
150 Views
Graphics provides one of the most natural means of communicating with a computer, since our highly developed 2D Or 3D pattern-recognition abilities allow us to perceive and process pictorial data rapidly. • Computers have become a powerful medium for the rapid and economical production of pictures. • Graphics provide a so natural means of communicating with the computer that they have become widespread. • Interactive graphics is the most important means of producing pictures since the invention of photography and television . • We can make pictures of not only the real world objects but als
- Categories:
 16 Views
16 Views
TRAINING AND PLACEMENT CELL is a web based application developed in the windows platform for the training and placement department of the college in order to provide the details of its students in a database for the companies to their process of recruitment provided with a proper login. The TRAINING AND PLACEMENT CELL contains all the information about the students. The system stores all the personal information of the students, like their personal details, their aggregate marks, their skill set and their technical skills that are required in the CV to be sent to a company.
- Categories:
 36 Views
36 Views
Tourism receipts worldwide are not expected to recover to 2019 levels until 2023. In
the first half of this year, tourist arrivals fell globally by more than 65 percent, with a near halt
since April—compared with 8 percent during the global financial crisis and 17 percent amid
the SARS epidemic of 2003, according to ongoing IMF research on tourism in a post-pandemic
world. Because of pandemic we faces the different struggles specially the business closed.
that’s why country’s economy decrease, at first many company need to reduce their employee.
- Categories:
 39 Views
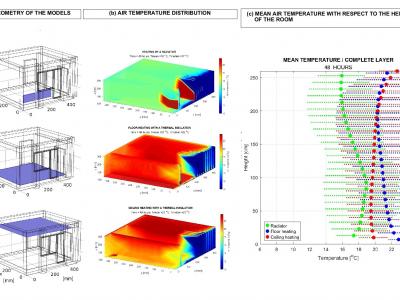
39 ViewsWith the development of recommender systems (RSs), several promising systems have emerged, such as context-aware RS, multi-criteria RS, and group RS. Multi-criteria recommender systems (MCRSs) are designed to provide personalized recommendations by considering user preferences in multiple attributes or criteria simultaneously. Unlike traditional RSs that typically focus on a single rating, these systems help users make more informed decisions by considering their diverse preferences and needs across various dimensions.
- Categories:
 155 Views
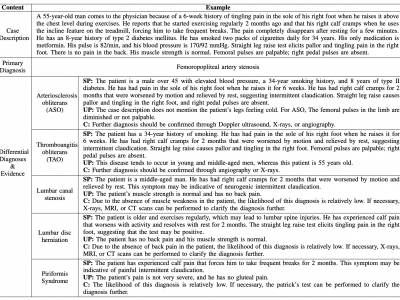
155 ViewsNumerical simulations are used to assess the efficiency of floor heating, ceiling heating, and plane radiator heating in a selected family house room under winter conditions in the Central European climate zone. COMSOL Multiphysics software was used for computer simulations. The output data were subsequently processed and analyzed using MATLAB software. Results indicate that floor and ceiling heating systems achieve higher and more rapid temperature increases compared to plane radiators.
- Categories:
 137 Views
137 Views
The data set includes attack implementations in an Internet of Things (IoT) context. The IoT nodes use Contiki-NG as their operating system and the data is collected from the Cooja simulation environment where a large number of network topologies are created. Blackhole and DIS-flooding attacks are implemented to attack the RPL routing protocol.
The datasets includes log file output from the Cooja simulator and a pre-processed feature set as input to an intrusion detection model.
- Categories:
 188 Views
188 Views
Offline-to-online is a key strategy for advancing reinforcement learning towards practical applications. This approach not only reduces the risks and costs associated with online exploration, but also accelerates the agent’s adaptation to real-world environments. It consists of two phases: offline-training and fine-tuning. However, offline-training and fine-tuning have different problems. In offline-training, the main difficulty is how to learn an excellent policy in a limited and incompletely distributed dataset.
- Categories:
 74 Views
74 Views