Signal Processing
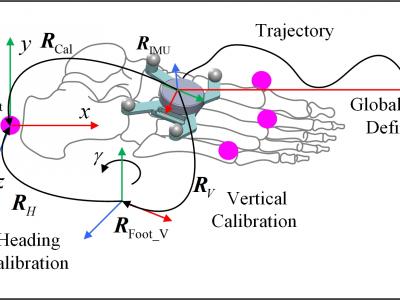
Knee osteoarthritis (KOA) is a common joint disease, causing pain, stiffness and other motor disorders. Gait analysis is an important basis for understanding the causes and formulating personalized rehabilitation plans. Although optical motion capture is regarded as the gold standard for measurement, it has drawbacks such as high cost and being limited to laboratories.
- Categories:
 83 Views
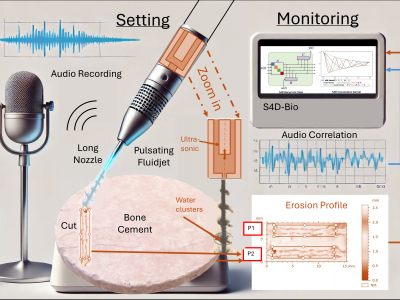
83 ViewsThis dataset comprises extensive multi-modal data related to the experimental study of ultrasonically excited pulsating fluid jets used for bone cement removal. Conducted at the Institute of Geonics, Ostrava, Czech Republic, the study explores the effect of varying standoff distances on erosion profiles, under controlled parameters including a fixed nozzle diameter, sonotrode frequency, supply pressure, and robot arm velocity. The dataset includes numerical data representing ablation profiles, captured as a large CSV file, and audio recordings captured using a high-resolution microphone.
- Categories:
 146 Views
146 Views
With the accelerating pace of population aging, the urgency and necessity for elderly individuals to control smart home systems have become increasingly evident. Smart homes not only enhance the independence of older adults, enabling them to complete daily activities more conveniently, but also ensure safety through health monitoring and emergency alert systems, thereby reducing the caregiving burden on families and society.
- Categories:
 103 Views
103 Views
A novel ultra-low-voltage (ULV) Dual-EdgeTriggered (DET) flip-flop based on the True-Single-PhaseClocking (TSPC) scheme is presented in this paper. Unlike Single-Edge-Triggering (SET), Dual-Edge-Triggering has the advantage of operating at the half-clock rate of the SET clock. We exploit the TSPC principle to achieve the best energy-efficient figures by reducing the overall clock load (only to 8 transistors) and register power while providing fully static, contention-free functionality to satisfy ULV operation.
- Categories:
 21 Views
21 ViewsJamming devices present a significant threat by disrupting signals from the global navigation satellite system (GNSS), compromising the robustness of accurate positioning. The detection of anomalies within frequency snapshots is crucial to counteract these interferences effectively. A critical preliminary measure involves the reliable classification of interferences and characterization and localization of jamming devices.
- Categories:
 166 Views
166 ViewsJamming devices pose a significant threat by disrupting signals from the global navigation satellite system (GNSS), compromising the robustness of accurate positioning. Detecting anomalies in frequency snapshots is crucial to counteract these interferences effectively. The ability to adapt to diverse, unseen interference characteristics is essential for ensuring the reliability of GNSS in real-world applications. We recorded a dataset with our own sensor station at a German highway with two interference classes and one non-interference class.
- Categories:
 238 Views
238 Views
This rolling bearing dataset was collected using the QPZZ-II rotating machinery fault simulation test rig developed by Jiangsu Qianpeng Diagnostic Engineering Co., Ltd. The bearing used in the experiment was model NU205EM. An electromyographic-type acceleration sensor was used in this experiment, which was placed in the horizontal X and vertical Y directions of the bearing housing on the test rig.
- Categories:
 77 Views
77 Views
A group of 10 healthy subjects without any upper limb pathologies participated in the data collection process. A total of 8 activities are performed by each subject. The measurement setup consists of a 5-channel Noraxon Ultium wireless sEMG sensor system. Representative muscle sites of the forearm are identified and self-adhesive Ag/AgCl dual electrodes are placed. The signal (sEMG) recorded during an ADL activity is segmented into functional phases: 1) rest 2) action and 3) release. During the rest phase, the subject is instructed to rest the muscles in a natural way.
- Categories:
 86 Views
86 Views
The data utilized the IWR1642 FMCW radar and the DCA1000EVM data acquisition board from Texas Instruments. Three different environments—bedroom, shared office, and unoccupied conference room—were selected as potential scenarios for non-contact vital sign monitoring. Continuous monitoring was conducted at distances of 0.5, 1, 1.5, and 2m for 120s. The results were calculated using a 30-second data window, with 91 calculations performed during each monitoring session, starting from the 30th second and continuing until the 120th second.
- Categories:
 61 Views
61 Views
This code accompanies the paper titled "An End-to-End Modular Framework for Radar Signal Processing: A Simulation-Based Tutorial," published in IEEE Aerospace and Electronics Systems Magazine (DOI:10.1109/MAES.2023.3334689). The simulation of a complete radar has been performed, and the results have been shown after each stage/module of a radar, enabling the reader to appreciate the impact of the specific process on the radar signal.
- Categories:
 156 Views
156 Views