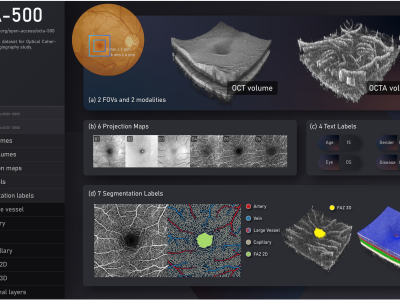
Demeaned images for SV2A ICA analysis

- Citation Author(s):
-
Daniele Bertoglio (University of Antwerp)Jordy Akkermans (University of Antwerp)
- Submitted by:
- Daniele Bertoglio
- Last updated:
- DOI:
- 10.21227/8jda-j668
 84 views
84 views
- Categories:
Abstract
Instructions:
Nifty files of each subject derived from the parametric maps are included as described in the readme.doc file.