Biophysiological Signals

8-channel monopolar sEMG signals were acquired using the device developed by our research group at a sampling rate of 1000 Hz. Medical gel electrodes (CH50B, Shanghai Hanjie Electronic Technology Co., LTD., Shanghai, China) were used for data collection. The position of the electrodes is shown in Fig. 2. The REF electrode was placed on the inner side of the upper big arm near the elbow and the RLD electrode was placed on the outer side of the right upper arm near the elbow. Eight monopolar electrodes were placed on the right forearm.
- Categories:
 125 Views
125 Views
This dataset is used to verify the effectiveness of the proposed MS-caCOH method.
- Categories:
 153 Views
153 ViewsThe large number and scale of natural and man-made disasters have led to an urgent demand for technologies that enhance the safety and efficiency of search and rescue teams. Semi-autonomous rescue robots are beneficial, especially when searching inaccessible terrains, or dangerous environments, such as collapsed infrastructures. For search and rescue missions in degraded visual conditions or non-line of sight scenarios, radar-based approaches may contribute to acquire valuable, and otherwise unavailable information.
- Categories:
 3576 Views
3576 Views
This dataset is associated with the manuscript entitled "Data-efficient Human Walking Speed Intent Inference". The data represent the measurements taken from 15 able-bodied human subjects as the made speed changes while walking on a treadmill. Each subject is associated with a .mat file that contains 8 variables. Four variables are associated with the training dataset while four are associated with the experimental testing protocol.
- Categories:
 123 Views
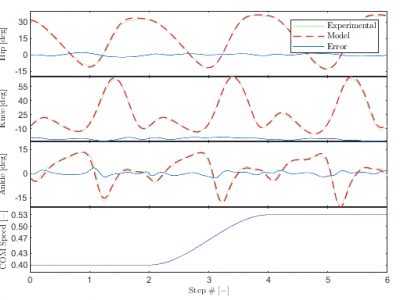
123 ViewsData and code associated with the submitted paper "A Kinematic Model to Predict a Continuous Range of Human-like Walking Speed Transitions" by Greggory F. Murray and Anne E. Martin.
This submission contains the experimental kinematic data for the speed transitions and the code needed to analyze and model the transitions.
- Categories:
 154 Views
154 Views
10 soccer supporters gathered to watch a live broadcasted Premier League
match between Liverpool and Manchester United (4 - 0) on 19th of March 2022, all
equipped with wrist-worn accelerometers. All participants were aware of the purpose of this experiment and consented to participate
by attendance at the event, and by wearing the accelerometer. No personally sensitive
information was collected, all data is fully anonymised following the GDPR guidelines
and all procedures were in accordance with the recommendations of the data protection
- Categories:
 150 Views
150 ViewsIREYE4TASK is a dataset for wearable eye landmark detection and mental state analysis. Sensing the mental state induced by different task contexts, where cognition is a focus, is as important as sensing the affective state where emotion is induced in the foreground of consciousness, because completing tasks is part of every waking moment of life. However, few datasets are publicly available to advance mental state analysis, especially those using the eye as the sensing modality with detailed ground truth for eye behaviors.
- Categories:
 1030 Views
1030 ViewsDesign of EEG-TMS experiment. The figure shows the timeline of the experimental session, the illustration of the typical sequence of the visual cues and the structure of one trial, and the illustration of the time intervals of interest within a trial: Pre is the baseline pre-que interval [-4.5 -0.5] s; Post is the post-cue interval [0 0.5] s; Img is the interval [1 3] s of motor imagery execution; here, t=0 corresponds to the moment of the appearance of the visual cue to start the movement.
- Categories:
 206 Views
206 ViewsWearable long-term monitoring applications are becoming more and more popular in both the consumer and the medical market. In wearable ECG monitoring, the data quality depends on the properties of the electrodes and on how they contact the skin. Dry electrodes do not require any action from the user. They usually do not irritate the skin, and they provide sufficiently high-quality data for ECG monitoring purposes during low-intensity user activity. We investigated prospective motion artifact–resistant dry electrode materials for wearable ECG monitoring.
- Categories:
 158 Views
158 Views