Biophysiological Signals
With the continuous advancement of technology, small and portable physiological sensors that can be worn on the body are quietly integrating into our daily lives, and are expected to greatly enhance our quality of life. In order to further enrich and expand the emotional physiological signals captured by portable wearable devices, we utilized the 14-channel portable EEG acquisition device Emotiv EPOC X, and with emotional video clips as the stimulus source, we collected two sets of emotional EEG signals from two groups of 10 participants each, named EmoX1 and EmoX2.
- Categories:
 273 Views
273 ViewsStress became a common factor of individuals in this competitive work environment, especially in academics. To address and assess this issue, this MUSEI-EEG dataset provides the Electroencephalogram (EEG) data of 20 undergraduate individuals in the 18-24 years age group (both male and female). Raag Darbari's music-based three-stage paradigm is designed for the subjects for cognitive stress assessment. Through this paradigm, physiological signal-based monitoring of stress level reduction can be observed in reference to stress and anxiety forms filled by the individual.
- Categories:
 457 Views
457 Views
This study introduces a multimodal dataset collected to investigate the psychophysiological relationship between perceived fear and muscle activity in climbers. The dataset includes physiological, motion, and subjective data from 19 climbers during \textit{lead} and \textit{top rope climbing} ascent styles, which differ in perceived risk due to varying fall distances. Physiological data were recorded using EMG, ECG, and IMU sensors. Additionally, subjective fear ratings were collected at distinct phases of the climb.
- Categories:
 93 Views
93 Views
Two publicly available datasets, the PASS and EmpaticaE4Stress databases, were utilised in this study. They were chosen because they both used the same Empatica E4 device, which allowed the acquisition of a variety of signals, including PPG and EDA. The dataset consists of in 1587 30-second PPG segments. Each segment has been filtered and normalized using a 0.9–5 Hz band-pass and min-max normalization scheme.
- Categories:
 170 Views
170 Views
A group of 10 healthy subjects without any upper limb pathologies participated in the data collection process. A total of 8 activities are performed by each subject. The measurement setup consists of a 5-channel Noraxon Ultium wireless sEMG sensor system. Representative muscle sites of the forearm are identified and self-adhesive Ag/AgCl dual electrodes are placed. The signal (sEMG) recorded during an ADL activity is segmented into functional phases: 1) rest 2) action and 3) release. During the rest phase, the subject is instructed to rest the muscles in a natural way.
- Categories:
 159 Views
159 ViewsBrain-Computer Interface (BCI) technology facilitates a direct connection between the brain and external devices by interpreting neural signals. It is critical to have datasets that contain patient's native languages while developing BCI-based solutions for neurological disorders. However, present BCI research lacks appropriate language-specific datasets, particularly for languages such as Telugu, which is spoken by more than 90 million people in India.
- Categories:
 434 Views
434 Views
This dataset comprises radar-acquired signals from 15 subjects walking on a treadmill, aimed at exploring methodologies for non-contact vital sign detection under conditions of significant body movement. Each subject participated in four experimental sessions, where radar data were collected using two Continuous Wave (CW) radars positioned to capture signals from the front and back of the subject. The data includes both raw and demodulated signals synchronized with ground-truth data obtained from a BioPac system.
- Categories:
 132 Views
132 Views
In this study, we collected EEG and EMG data from 16 subjects during the MI process and constructed a homemade MI-hBCI dataset. The participants included 10 males (mean age: 22.3±3.1 years) and 6 females (mean age: 22.1±2.4 years). All the subjects were right-handed, had normal vision, and had no motor impairment; all the participants signed a consent form and were informed of the experimental procedure and precautions before the experiment.
- Categories:
 193 Views
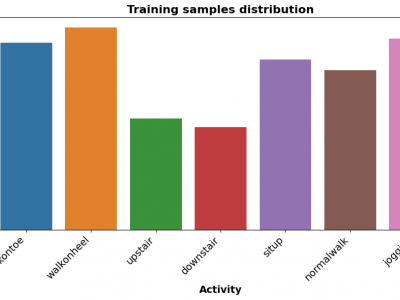
193 ViewsThe Human Activity Recognition (HAR) dataset comprises comprehensive data collected from various human activities including walking, running, sitting, standing, and jumping. The dataset is designed to facilitate research in the field of activity recognition using machine learning and deep learning techniques. Each activity is captured through multiple sensors providing detailed temporal and spatial data points, enabling robust analysis and model training.
- Categories:
 254 Views
254 Views