Biophysiological Signals

In this study, we collected EEG and EMG data from 16 subjects during the MI process and constructed a homemade MI-hBCI dataset. The participants included 10 males (mean age: 22.3±3.1 years) and 6 females (mean age: 22.1±2.4 years). All the subjects were right-handed, had normal vision, and had no motor impairment; all the participants signed a consent form and were informed of the experimental procedure and precautions before the experiment.
- Categories:
 194 Views
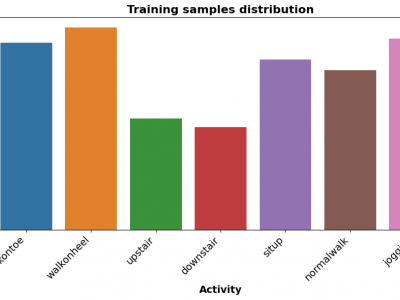
194 ViewsThe Human Activity Recognition (HAR) dataset comprises comprehensive data collected from various human activities including walking, running, sitting, standing, and jumping. The dataset is designed to facilitate research in the field of activity recognition using machine learning and deep learning techniques. Each activity is captured through multiple sensors providing detailed temporal and spatial data points, enabling robust analysis and model training.
- Categories:
 260 Views
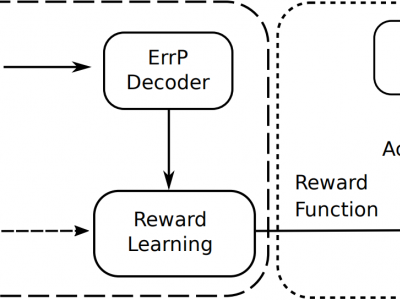
260 ViewsDeveloping mind-controlled prosthetics that seamlessly integrate with the human nervous system is a significant challenge in the field of bioengineering. This project investigates the use of labelled brainwave patterns to control a bionic arm equipped with a sense of touch. The core objective is to establish a communication channel between the brain and the artificial limb, enabling intuitive and natural control while incorporating sensory feedback.
The project involves:
- Categories:
 741 Views
741 Views
The dataset consists of 4-channeled EOG data recorded in two environments. First category of data were recorded from 21 poeple using driving simulator (1976 samples). The second category of data were recorded from 30 people in real-road conditions (390 samples).
All the signals were acquired with JINS MEME ES_R smart glasses equipped with 3-point EOG sensor. Sampling frequency is 200 Hz.
- Categories:
 178 Views
178 Views
The dataset involves two sets of participants: a group of twenty skilled drivers aged between 40 and 68, each having a minimum of ten years of driving experience (class 1), and another group consisting of ten novice drivers aged between 18 and 46, who were currently undergoing driving lessons at a driving school (class 2).
The data was recorded using JINS MEME ES_R smart glasses by JINS, Inc. (Tokyo, Japan).
Each file consists of a signals from one sigle ride.
- Categories:
 78 Views
78 Views
The AnxiECG-PPG Database contains synchronized electrocardiogram (ECG) and mobile-acquired photoplethysmography (PPG) recordings from 47 healthy participants. Moreover, the acquisition protocol assesses three distinct states: a 5-minute Baseline, a 1-minute Physical Activated State, and a Psychological Activated state provoked through emotion-induced videos (negative, positive, and neutral emotion valence).
- Categories:
 1580 Views
1580 Views
Normal
0
false
false
false
EN-US
X-NONE
AR-SA
- Categories:
 214 Views
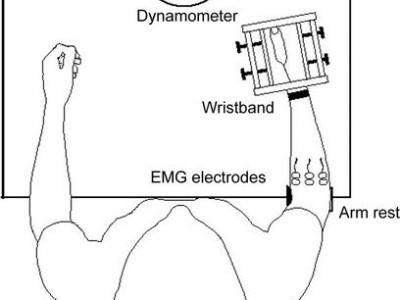
214 ViewsThis data was recorded for emg based force/Torque estimation. EMG and torque signals were collected during simultaneous, isometric, but continuously varying contractions, corresponding to two wrist DoF. The experiment was carried out in two trials with a 5-min rest in between. Each trial included six combinations of tasks, separated by 2 min of rest to minimize the effect of fatigue. The performed tasks were categorized into individual and combined (simultaneous) DoF to test the ability to estimate isolated torque and torque in two simultaneous DoF.
- Categories:
 287 Views
287 Views