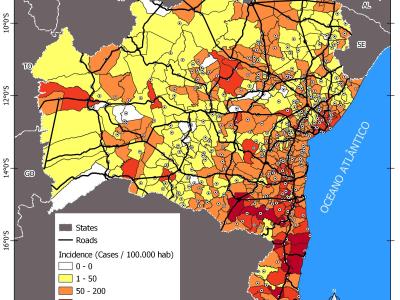
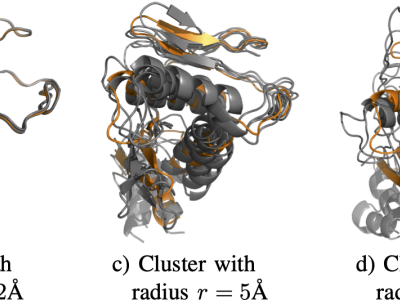
In My study, we evaluate the performance of the proposed clustering method across a wide range of publicly available datasets that represent different data modalities. Specifically, Jaffe, ExtendYaleB, and ORL are employed as facial image datasets to assess the method's capability in handling variations in facial expressions and lighting conditions.
- Categories: