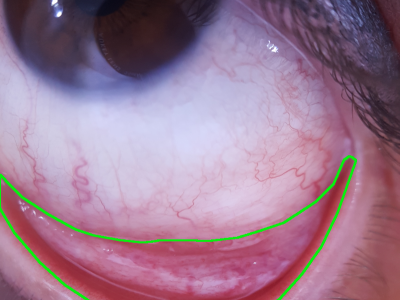
To investigate SAM's potential in the continual scenario, we construct a benchmark for continual segmentation, called Continual SAM Adaptation Benchmark (CoSAM), which aims to systematically evaluate SAM-related algorithms's performance in the streaming scenarios. Specifically, CoSAM offers a set of 8 tasks covering diverse domains, including industrial defects, medical imaging, and camouflaged objects, to serve as a realistic and effective benchmark for evaluating current methods.
- Categories: