Electroencephalography
Synthetic Epileptic Spike EEG Database (SESED-WUT)
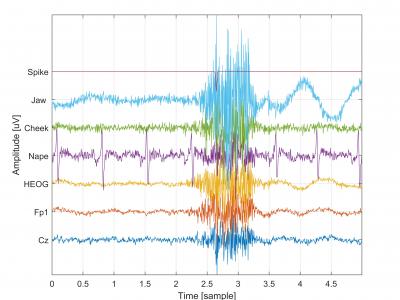
The database contains EEG, EMG, and EOG signals with artificially generated epileptic spikes. The recordings were performed using the g.USBamp 2.0 amplifier. Data were collected from 5 EEG channels (C3, Cz, C4, Fz, Fp1), 1 EOG channel (VEOG), and 3 EMG channels (Nape, Cheek, Jaw). The signals were sampled at 256 Hz and processed with a bandpass filter (0.1–100 Hz) and a notch filter (48–52 Hz).
- Categories:
 292 Views
292 ViewsTen volunteers were trained through a series of twelve daily lessons to type in a computer using the Colemak keyboard layout. During the fourth-, eight-, and eleventh-session, electroencephalography (EEG) measurements were acquired for the five trials each subject performed in the corresponding lesson. Electrocardiography (ECG) data at each of those trials were acquired as well. The purpose of this experiment is to aim in the development of different methods to assess the process of learning a new task.
- Categories:
 1316 Views
1316 ViewsDataset description
This dataset contains EEG signals from 73 subjects (42 healthy; 31 disabled) using an ERP-based speller to control different brain-computer interface (BCI) applications. The demographics of the dataset can be found in info.txt. Additionally, you will find the results of the original study broken down by subject, the code to build the deep-learning models used in [1] (i.e., EEG-Inception, EEGNet, DeepConvNet, CNN-BLSTM) and a script to load the dataset.
Original article:
- Categories:
 3158 Views
3158 Views
Ear-EEG recording collects brain signals from electrodes placed in the ear canal. Compared with existing scalp-EEG, ear-EEG is more wearable and user-comfortable compared with existing scalp-EEG.
- Categories:
 2592 Views
2592 ViewsOur state of arousal can significantly affect our ability to make optimal decisions, judgments, and actions in real-world dynamic environments. The Yerkes-Dodson law, which posits an inverse-U relationship between arousal and task performance, suggests that there is a state of arousal that is optimal for behavioral performance in a given task. Here we show that we can use on-line neurofeedback to shift an individual's arousal from the right side of the Yerkes-Dodson curve to the left toward a state of improved performance.
- Categories:
 6524 Views
6524 Views
Previous neuroimaging research has been traditionally confined to strict laboratory environments due to the limits of technology. Only recently have more studies emerged exploring the use of mobile brain imaging outside the laboratory. This study uses electroencephalography (EEG) and signal processing techniques to provide new opportunities for studying mobile subjects moving outside of the laboratory and in real world settings. The purpose of this study was to document the current viability of using high density EEG for mobile brain imaging both indoors and outdoors.
- Categories:
 1197 Views
1197 ViewsElectroencephalography (EEG) signal data was collected from twelve healthy subjects with no known musculoskeletal or neurological deficits (mean age 25.5 ± 3.7, 11 male, 1 female, 1 left handed, 11 right handed) using an EGI Geodesics© Hydrocel EEG 64-Channel spongeless sensor net. All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the University of Wisconsin-Milwaukee (17.352).
- Categories:
 1445 Views
1445 ViewsRecent advances in scalp electroencephalography (EEG) as a neuroimaging tool have now allowed researchers to overcome technical challenges and movement restrictions typical in traditional neuroimaging studies. Fortunately, recent mobile EEG devices have enabled studies involving cognition and motor control in natural environments that require mobility, such as during art perception and production in a museum setting, and during locomotion tasks.
- Categories:
 4058 Views
4058 Views