GIB-UVa ERP-BCI dataset
- Citation Author(s):
- Submitted by:
- Eduardo Santamaria Vazquez
- Last updated:
- DOI:
- 10.21227/6bdr-4w65
- Data Format:
- Research Article Link:
 3170 views
3170 views
- Categories:
- Keywords:
Abstract
Dataset description
This dataset contains EEG signals from 73 subjects (42 healthy; 31 disabled) using an ERP-based speller to control different brain-computer interface (BCI) applications. The demographics of the dataset can be found in info.txt. Additionally, you will find the results of the original study broken down by subject, the code to build the deep-learning models used in [1] (i.e., EEG-Inception, EEGNet, DeepConvNet, CNN-BLSTM) and a script to load the dataset.
Original article:
[1] Santamaría-Vázquez, E., Martínez-Cagigal, V., Vaquerizo-Villar, F., & Hornero, R. (2020). EEG-Inception: A Novel Deep Convolutional Neural Network for Assistive ERP-based Brain-Computer Interfaces. IEEE Transactions on Neural Systems and Rehabilitation Engineering. https://doi.org/10.1109/TNSRE.2020.3048106
Some people report problems to register and use IEEE Dataport. Additional sources:
Official code repository: https://github.com/esantamariavazquez/EEG-Inception
Dataset copy in kaggle: https://www.kaggle.com/esantamaria/gibuva-erpbci-dataset
Instructions:
This dataset contains data from 73 subjects using an ERP-based speller with the row-column paradigm (RCP) as stimulation paradigm. You can find complete information about the experiments in [1].
The dataset contains the following variables:
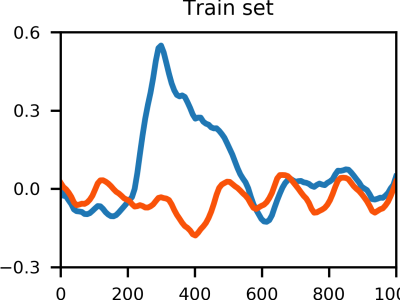
- features [n_stimuli x n_samples x n_channels] -> EEG epochs [0, 1000] ms after stimulus onset. The EEG is already preprocessed (i.e., FIR bandpass filter order 1000 [0.5-45] Hz, Common average reference (CAR), baseline normalization [-200, 0] ms). Channel order: ['FZ', 'CZ', 'PZ', 'P3', 'P4', 'PO7', 'PO8', 'OZ'].
- erp_labels [n_stimuli x 1] -> ERP labels (i.e., 0 for non-target stimulus, 1 for target stimulus)
- codes [n_stimuli x 1] -> Code of the row or column that was highlighted
- trials [n_stimuli x 1] -> Array that relates each stimulus to its trial. Use example: features[trials==10] returns the EEG epochs of the 10th trial.
- sequences [n_stimuli x 1] -> Array that relates each stimulus to its number of sequence. Use example: features[sequences<=10] returns the EEG epochs from sequences 1-10.
- subjects [n_stimuli x 1] -> Array that relates each stimulus to its subject. Use example: features[subjects==10] returns the EEG epochs of subject 10.
- database_ids [n_stimuli x 1] -> Array that relates each stimulus to its database (there are 3 different databases, as explained in the original study)
- run_indexes [n_stimuli x 1] -> Array that relates each stimulus to its run.
- matrix_indexes [n_stimuli x 1] -> Array that relates each stimulus to its matrix_index.
- target [n_trials x 1] -> Trial target. Column information [database_id, subject, trial, run, matrix_index, row, col]
- matrix_dims [n_runs x 1] -> Matrix dimmensions for each run
Original article:
Santamaría-Vázquez, E., Martínez-Cagigal, V., Vaquerizo-Villar, F., & Hornero, R. (2020). EEG-Inception: A Novel Deep Convolutional Neural Network for Assistive ERP-based Brain-Computer Interfaces. IEEE Transactions on Neural Systems and Rehabilitation Engineering. https://doi.org/10.1109/TNSRE.2020.3048106






In reply to This dataset contains EEG by BASAVARAJU C
In reply to What is the correspondence by Rajeev Sahay