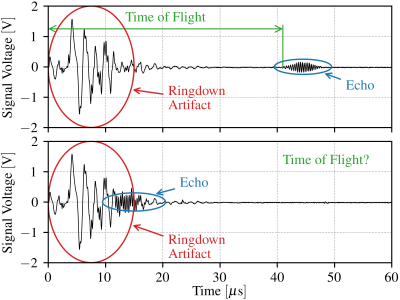
Ultrasound Waveforms with and without Ringdown Artifacts
- Citation Author(s):
- Submitted by:
- Yana Sosnovskaya
- Last updated:
- DOI:
- 10.21227/z6v5-mf23
- Data Format:
- Research Article Link:
 905 views
905 views
- Categories:
- Keywords:
Abstract
Minimally-Invasive Surgeries can benefit from having miniaturized sensors on surgical graspers to provide additional information to the surgeons. One such potential sensor is an ultrasound transducer. At long travel distances, the ultrasound transducer can accurately measure its ultrasound wave's time of flight, and from it, classify the grasped tissue. However, the ultrasound transducer has a ringing artifact arising from the decaying oscillation of its piezo element, and at short travel distances, the artifact blends with the acoustic echo. Without a method to remove the artifact from the blended signal, this makes it impossible to measure the waveform's time of flight.
It is possible to use both classical signal processing and deep learning methods to filter raw ultrasound signals, removing the ringing artifact from them, and from the filtered signals, to obtain the time of flight. In this dataset, two datasets are provided to train and test algorithms developed for filtering out the ringdown artifact, and for subsequently extracting the waveform's time of flight. All measured (raw) signals were collected the same experimental setup: an oscilloscope connected to an ultrasound driver to drive a transducer attached to a liquid water container, in an attempt to mimic tissue properties in a tightly controlled environment.
The training dataset consists of two groups of signal pairs. The first group consists of 993 signal pairs, with each pair consisting of a raw ultrasound signal (with the acoustic echo blended with the ringing artifact), and a target filtered signal (with only the desired echo). Signals in the first group are sampled at the original sampling frequency of 500 MHz. The second group is like the first group, but with all signals downsampled by a factor of 26. This training dataset includes only travel distances from 2 cm to 4 cm, inclusively, because at these distances in water, the echo is sufficiently separated from the ringdown artifact to be manually extractable. The signal pairs are approximately equally distributed between the distances covered.
The test dataset similarly consists of two groups of raw ultrasound signals. The first group consists of 270 signals, collected at 9 travel distances between 0.5 cm and 4.0 cm, with 30 signals per distance. It also includes the associated true times of flight for each distance. Signals in the first group are sampled at the original sampling frequency of 500 MHz. The second group is like the first group, but with all signals downsampled by a factor of 26. All signals in both datasets are aligned.
Instructions:
In both attached datasets, the signals are in Volts, the time values are in microseconds, and the distances are in cm. Both files are MATLAB .mat files, that will require at minimum MATLAB v7.3 (R2006b) or later to load. NumPy 1.20.1 works as well, as will likely much older versions.
The training dataset 'TrainDataset.mat' can be read via the provided Python script ReadTrainData.py. Refer to the dataset abstract for details about how the training data was collected. All waveforms are aligned. 'TrainDataset.mat' contains the following variables:
- 'Data_in': 993 raw ultrasound waveforms, at the original sampling frequency of 500 MHz;
- 'Data_out': same waveforms as in Data_in, with the ringing artifacts manually zeroed, also at the original sampling frequency of 500 MHz;
- 'TimeVector': time vector of the waveforms in Data_in and Data_out;
- 'Data_in_downsampled_26': Data_in downsampled by a factor of 26;
- 'Data_out_downsampled_26': Data_out downsampled by a factor of 26;
- 'TimeVector_downsampled_26': TimeVector downsampled by a factor of 26.
The test dataset 'TestDataset.mat' can be read via the provided Python script ReadTestData.py. Refer to the dataset abstract for details about how the test data was collected. All waveforms are aligned. 'TestDataset.mat' contains the following variables:
- 'Distance': distances that were used for data collection, in descending order;
- 'TOF': times of flight calculated based on the distance and the velocity of sound in liquid water at room temperature and pressure, in descending order;
- 'Test_data_XXmm': 30 raw ultrasound waveforms for the travel distance of XX mm. The travel distances covered are the same as in Distance:
- 40 mm
- 35 mm
- 30 mm
- 25 mm
- 20 mm
- 15 mm
- 10 mm
- 08 mm
- 05 mm
- 'TimeVector': time vector for the above test waveforms.
- 'Test_data_downsampled_26_XXmm': 30 raw ultrasound waveforms for the travel distance of XX mm, downsampled by a factor of 26. The travel distances covered are the same as in Distance:
- 40 mm
- 35 mm
- 30 mm
- 25 mm
- 20 mm
- 15 mm
- 10 mm
- 08 mm
- 05 mm
- 'TimeVector_downsampled_26': time vector of the above test waveforms, downsampled by a factor of 26.