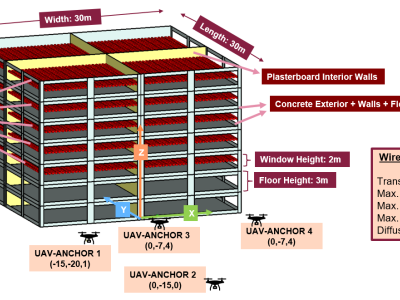
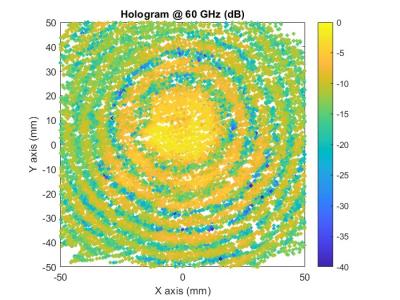
These are three sets of simulated echo data generated by Matlab. The first set of echo data comes from a surface target with a size comparable to a 2-D MIMO array, but with a center point offset from the array. The second group contains three echo data from point targets. The first packet is a point target within the physical size of the array, the second packet is a point target outside the physical size of the array, and the third packet is the sum of the first two packets.
- Categories: