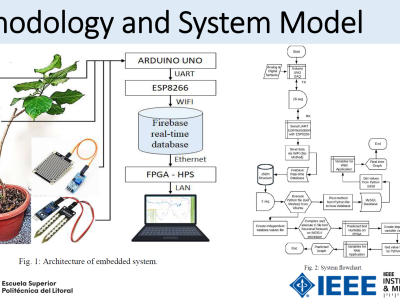
UniTOBrain
- Citation Author(s):
-
Edwin Bennink
(Department of Radiology, University Medical Center Utrecht, Utrecht, the Netherlands, Image Sciences Institute, University Medical Center Utrecht, Utrecht, the Netherlands)
Francesca Bertolino (Neurosciences Department, University of Turin (Italy)) - Submitted by:
- apns apns
- Last updated:
- DOI:
- 10.21227/x8ea-vh16
- Data Format:
 4043 views
4043 views
- Categories:
- Keywords:
Abstract
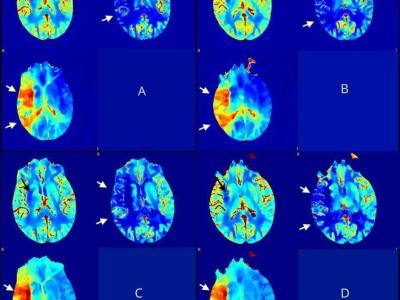
The University of Turin (UniTO) released the open-access dataset Stoke collected for the homonymous Use Case 3 in the DeepHealth project (https://deephealth-project.eu/). UniToBrain is a dataset of Computed Tomography (CT) perfusion images (CTP). The dataset includes 258 consecutive patients, a subsample of 100 training subjects and 15 testing subjects was used in a submitted publication for the training and the testing of a Convolutional Neural Network (CNN, see for details: https://arxiv.org/abs/2101.05992, https://paperswithcode.com/paper/neural-network-derived-perfusion-maps-a-model, https://www.medrxiv.org/content/10.1101/2021.01.13.21249757v1). The UniTO team released this dataset publicly.
CTP data were retrospectively obtained from the hospital PACS of Città della Salute e della Scienza di Torino (Molinette). CTP acquisition parameters were as follows: Scanner GE, 64 slices, 80 kV, 150 mAs, 44.5 sec duration, 89 volumes (40 mm axial coverage), injection of 40 ml of Iodine contrast agent (300 mg/ml) at 4 ml/s speed.
Along with the dataset, we provide some utility files.
dicomtonpy.py: It converts the dicom files in the dataset to numpy arrays. These are 3D arrays, where CT slices at the same height are piled-up over the temporal acquisition.
dataloader_pytorch.py: Dataloader for the pytorch deep learning framework. It converts the numpy arrays in normalized tensors, which can be provided as input to standard deep learning models.
dataloader_pyeddl.py: Dataloader for the pyeddl deep learning framework. It converts the numpy arrays in normalized tensors, which can be provided as input to standard deep learning models using the european library EDDL. Visit https://github.com/EIDOSlab/UC3-UNITOBrain to have a full companion code where a U-Net model is trained over the dataset.
Instructions:
Visit https://github.com/EIDOSlab/UC3-UNITOBrain to have a full companion code where a U-Net model is trained over the dataset.