Datasets
Standard Dataset
Ultrasound spinal cord dataset
- Citation Author(s):
- Submitted by:
- Avisha Kumar
- Last updated:
- Fri, 01/03/2025 - 09:55
- DOI:
- 10.21227/33h3-qx42
- Data Format:
- License:
 154 Views
154 Views- Categories:
- Keywords:
Abstract
While deep learning has catalyzed breakthroughs across numerous domains, its broader adoption in clinical settings is inhibited by the costly and time-intensive nature of data acquisition and annotation. To further facilitate medical machine learning, we present an ultrasound dataset of 10,223 Brightness-mode (B-mode) images consisting of sagittal slices of porcine spinal cords (N=25) before and after a contusion injury. We additionally benchmark the performance metrics of several state-of-the-art object detection algorithms to localize the site of injury and semantic segmentation models to label the anatomy for comparison and creation of task-specific architectures. Finally, we evaluate the zero-shot generalization capabilities of the segmentation models on human ultrasound spinal cord images to determine whether training on our porcine dataset is sufficient for accurately interpreting human data. Our results show that the YOLOv8 detection model outperforms all evaluated models for injury localization, achieving a mean Average Precision (mAP50-95) score of 0.606. Segmentation metrics indicate that the DeepLabv3 segmentation model achieves the highest accuracy on unseen porcine anatomy, with a Mean Dice score of 0.587, while SAMed achieves the highest Mean Dice score generalizing to human anatomy (0.445). To the best of our knowledge, this is the largest annotated dataset of spinal cord ultrasound images made publicly available to researchers and medical professionals, as well as the first public report of object detection and segmentation architectures to assess anatomical markers in the spinal cord for methodology development and clinical applications.
Dataset from manuscript "A novel open-source ultrasound dataset with deep learning benchmarks for spinal cord injury localization and anatomical segmentation"
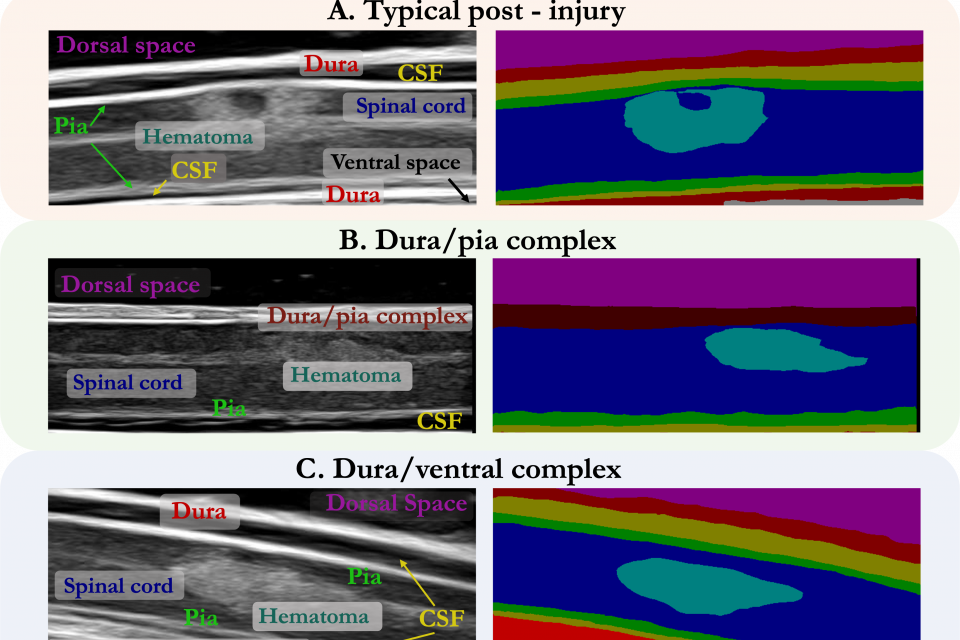
This is a dataset of 10,223 B-mode ultrasound images of porcine spinal cord before and after contracting an injury. The images were collected over 25 pigs at with varying aquisition parameters, such as spinal cord depth, imaging angle, gain, brightness, and ultrasound signal coupling medium.
Injury Localization
The injury localization dataset can also be accessed at this google drive link: https://drive.google.com/file/d/1r-r0dcXePMyXiXuVEUhtUgARw-Ktp_O1/view?usp=sharing
The images and their corresponding bounding box labels (saved as xml files) have been split into train, validation, and test folders. The injury localization dataset consists of 2,245 healthy and injured porcine spinal cord images (N=23). Images are in Portable Network Graphics (PNG) format and are cropped to size 690 x 275 pixels, which shows approximately 25 mm x 8 mm of the sagittal slice of the spinal cord.
Porcine Object Detection Dataset
Total images: 2,245
Total pre-injury images: 877
Total pre-injury pigs: 15
Total post-injury images: 1,368
Total post-injury pigs: 23
Semantic Segmentation
The segmentation dataset can also be accessed at this google drive link: https://drive.google.com/file/d/1JUKakYg10ZsHVDORsv9XvDkx86WVKaFO/view?usp=sharing
The semantic segmentation dataset consists of 10,223 images divided into 4,467 healthy spinal cord images (N=20) and 5,756 images of injured spinal cord images (N=25). Again, the files are in a PNG format, scaled to the depth of scan field on the ultrasound image, and cropped to size 690 x 275.
Porcine Segmentation Dataset
Total images: 10,223
Total pre-injury images: 4,467
Total pre-injury pigs: 20
Total post-injury images: 5,756
Total post-injury pigs: 25
Note: This work is under review.
Additional details about this dataset and the benchmarking analysis for injury localization and spinal cord segmentation can be found in our manuscript, which is currently under review.
Abstract: While deep learning has catalyzed break-throughs across numerous domains, its broader adoption in clinical settings is inhibited by the costly and time-intensive nature of data acquisition and annotation. To further facilitate medical computer vision, we present an ultrasound dataset of 10,223 Brightness-mode (B-mode) images consisting of sagittal slices of porcine spinal cords (N=25) before and after a contusion injury. We additionally benchmark the performance metrics of several state-of-the- art object detection algorithms to localize the site of injury and semantic segmentation models to label the anatomy for comparison and creation of task-specific architectures. Finally, we evaluate the zero-shot generalization capabilities of the segmentation models on human ultrasound spinal cord images to determine whether training on our porcine dataset is sufficient for these models to accurately interpret human data for clinical translation. Our results show that the YOLOv8 detection model outperforms all evaluated models for injury localization, achieving a mean Average Precision (mAP50-95) score of 0.606. Segmentation results indicate that the DeepLabv3 segmentation model achieves the highest accuracy on unseen porcine anatomy, with a Mean Dice score of 0.587, while SAMed achieves the highest Mean Dice score with unseen human anatomy (0.445). To the best of our knowledge, this is the largest annotated dataset of spinal cord ultrasound images made publicly available to researchers and medical professionals, as well as the first public report of object detection and segmentation architectures to assess anatomical markers in the spinal cord for methodology development and clinical applications.
Corresponding author: Avisha Kumar, PhD Student at Johns Hopkins University (akumar80 <@> jh <.> edu)
More from this Author
Dataset Files
- This zip files contains the injury localization dataset of ultrasound images of the porcine spinal cord and annotations. Injury localization dataset.zip (213.89 MB)
- This zip files contains the semantic segmentation dataset of ultrasound images of the porcine spinal cord and annotations. segmentation dataset (2).zip (1,019.65 MB)