Large-scale Genetic Exploration of Genome-wide Models: an Application to Human Cancer Metabolism
- Citation Author(s):
-
Youssef HamadiClaudio AngioneHillel KuglerChristoph WintersteigerBoyan Yordanov
- Submitted by:
- Youssef Hamadi
- Last updated:
- DOI:
- 10.21227/yegx-4z80
- Data Format:
 479 views
479 views
- Categories:
- Keywords:
Abstract
Computational modelling of metabolic processes has proven to be a useful approach to formulate our knowledge and improve our understanding of core biochemical systems that are crucial to maintain cellular functions. Recently, it has become evident that metabolism is not only responsible for generating the required energy and controlling the abundance of metabolites within a cell, but also has an important role in and influence on cellular fate specification. Toward understanding the broader role of metabolism on cellular decision-making in healthy and disease conditions, it is important to integrate the study of metabolism with other core regulatory systems within the cell, including gene regulatory networks that control gene expression patterns. However, the sheer size of human metabolism models makes large-scale integration and discovery difficult to perform extensively.
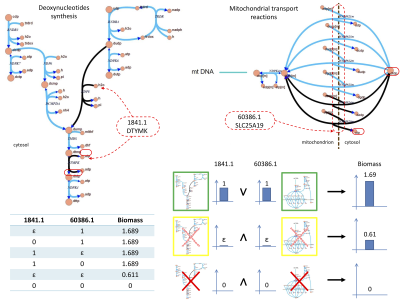
After quantitatively integrating gene expression profiles with a genome-scale reconstruction of human metabolism, we propose combinatorial methods to reverse engineer gene expression profiles and combinations of genetic modifications that simultaneously optimize multi-objective cellular goals. This enables us to suggest classes of transcriptomic profiles that are most suitable to achieve given metabolic phenotypes. As a result, we can express objectives on the phenotypes and characterize genotypes able to reach the set objectives. We demonstrate how our techniques are able to compute beneficial, neutral or ``toxic'' combinations of gene expression levels. We test our method on a manually curated improved version of the human metabolic reconstruction (Recon 2), and on nine cell-specific cancer models, comparing our outcomes with the corresponding normal cells. When applied to cancer models, toxic genes become simultaneous targets for potential therapies.
Our methods enable large scale studies of metabolic reconstructions coupled to gene expression models, and open the way to a broad class of applications that require understanding of the interplay among genotype, metabolism, and cellular behavior. The code is freely available as a MATLAB toolbox.
Instructions:
The files presented here are represent extra experimental data and analysis for the aforementioned research paper.