STIR: Surgical Tattoos in Infrared
- Citation Author(s):
-
Omid Mohareri (Intuitive Surgical)Simon DiMaio (Intuitive Surgical)Septimiu Salcudean (The University of British Columbia)
- Submitted by:
- Dale Bergman
- Last updated:
- DOI:
- 10.21227/w8g4-g548
 2571 views
2571 views
- Categories:
- Keywords:
Abstract
Quantifying performance of methods for tracking and mapping tissue in endoscopic environments is essential for enabling image guidance and automation of medical interventions and surgery. Datasets developed so far either use rigid environments, visible markers, or require annotators to label salient points in videos after collection. These are respectively: not general, visible to algorithms, or costly and error-prone. We introduce a novel labeling methodology along with a dataset that uses said methodology, Surgical Tattoos in Infrared (STIR). STIR has labels that are persistent but invisible to visible spectrum algorithms. This is done by labelling tissue points with IR-flourescent dye, indocyanine green (ICG). STIR comprises hundreds of stereo video clips in both in vivo and ex vivo scenes with start and end points labelled in the IR spectrum. With over 3,000 labelled points, STIR will help to quantify and enable better analysis of tracking and mapping methods.
Instructions:
STIR: Surgical Tattoos in Infrared
# Dataset Paper
https://arxiv.org/abs/2309.16782
# Dataset format
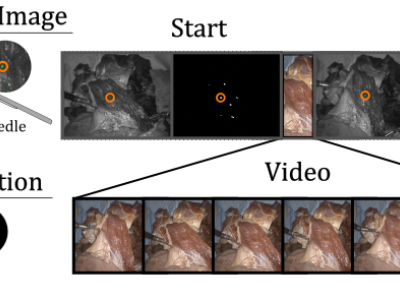
The dataset includes a set of numbered folders. Under each folder are
- left (left sequence folder)
- right (right sequence folder)
- calib.json (OpenCV camera calibration parameters for all subfolders)
Under each of left and right are:
seq** eg seq00-seq14 for example. Each 'seq' folder corresponds to an action.
For example, left/seq01 corresponds to the same recording as right/seq01 for the right camera. All images and videos are stereo rectified.
Each 'seq' folder has:
- frames (contains visible light video)
- segmentation (contains start and end binary segmentation pngs)
- icgstartseg.png (start segmentation)
- icgendseg.png (finish segmentation)
- <STARTTIME>ms_icgstart.png (starting firefly image)
- <STARTTIME>ms_icgend.png (finishing firefly image)
- icgbothimbox.png (frame with bounding boxes shown)
# Quantification
Only the icgstartseg, icgendseg, and frames video are needed for quantifying 2D performance. For evaluating in 3D, the calib.json file is necessary. Record using the video in the frames file at locations from icgstartseg.png, and then compare to the points in icgendseg using losses described in the paper (eg. chamfer/endpoint).
# Terms
By using the dataset, you agree to cite:
Schmidt, A., Mohareri, O., DiMaio, S., and Septimiu, S. E. Surgical Tattoos in Infrared: A Dataset for
Quantifying Tissue Tracking and Mapping. https://arxiv.org/abs/2309.16782
in any publication resulting from its use
# Contact information
Contact Adam Schmidt (adamschmidt@ece.ubc.ca) for any questions.







