NucExM Dataset for Nuclei Segmentation in Expansion Microscopy
- Citation Author(s):
-
Leander LauenburgZudi LinRuihan ZhangMarcia dos SantosSiyu HuangIgnacio Arganda-CarrerasEdward BoydenHanspeter PfisterDonglai Wei
- Submitted by:
- Zudi Lin
- Last updated:
- DOI:
- 10.21227/48b7-s821
- Data Format:
 363 views
363 views
- Categories:
- Keywords:
Abstract
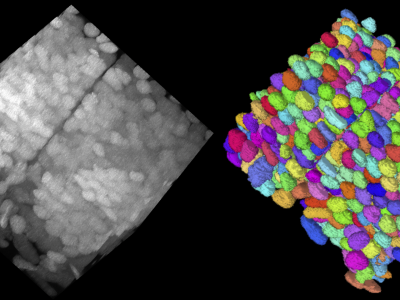
We curated the saturated nuclei segmentation annotation for two expansion microscopy (ExM) volumes by two neuroscience experts from a day 7 post-fertilization (dpf) zebrafish brain, imaged with confocal microscopy. These volumes have an anisotropic resolution of 0.325×0.325×2.5 μm in (x, y, z) order, with an approximate tissue expansion factor of 7.0. Thus the effective resolution becomes 0.046 × 0.046 × 0.357 μm. The two volumes are of size 2048×2048×255 voxels with 9.6K and 8.8K nuclei, respectively. This dataset is named the NucExM dataset, which serves as a testbed for 3D microscopy instance segmentation systems.
Instructions:
The dataset contains four volumes: two ExM image volumes and two 3D instance segmentation volumes.