Datasets
Standard Dataset
BEATing CILIAted cells
- Citation Author(s):
- Submitted by:
- Giovanni Dimauro
- Last updated:
- Fri, 12/15/2023 - 03:42
- DOI:
- 10.21227/qkm1-k152
- Data Format:
- Links:
- License:
 275 Views
275 Views- Categories:
- Keywords:
Abstract
Nasal cytology is a medicine field that focuses on the examination of nasal mucosa cells with the objective of recognizing changes in the epithelium, which is frequently subjected to acute or chronic irritation and inflammation caused by viruses, bacteria, or fungi; in the last decade, nasal cytology is becoming increasingly critical in diagnosing nasal conditions.
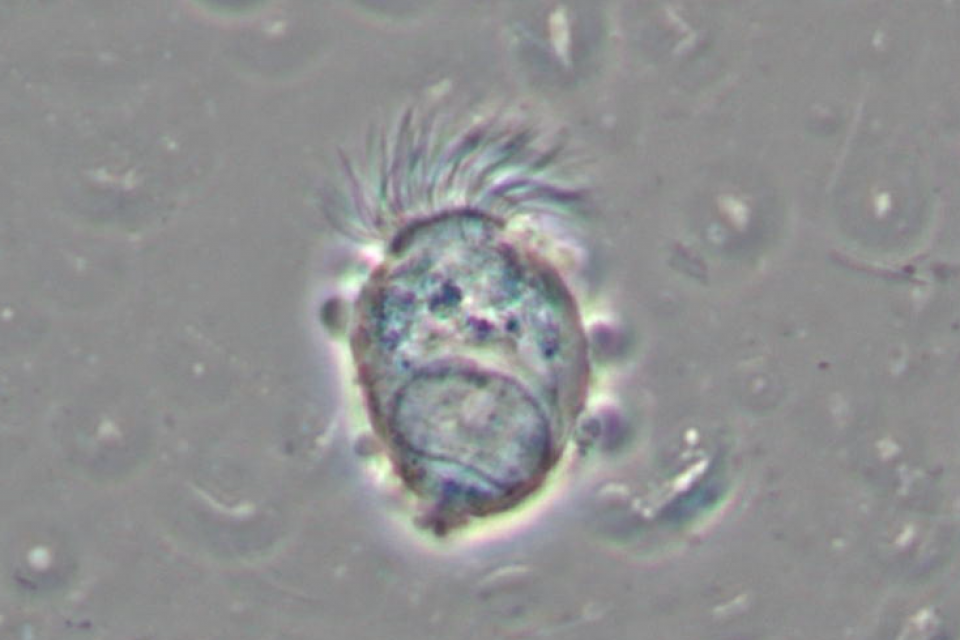
Among the cell population of the nasal mucosa, ciliated cells make up 80% of the epithelium in the upper airways. These cells are characterized by the presence of cilia that normally beat in a consistent, fluid pattern, easing effective mucociliary clearance.
This cytotype is quite important since its study allows also for the diagnosis of primary ciliary dyskinesia (PCD), a rare pathology that is usually associated with other severe pathologies such as situs inversus, cardiac diseases, and male infertility.
The assessment of ciliary function is done through the measurement of the ciliary beating frequency (CBF).
In addition, the CBF decay of the ciliated cells in a dead body may help to trace the time of death.
These motivations have fueled research seeking an architecture that supports such long, complex estimations, and feels necessary in medical and forensics contexts, that’s why we developed Deep Cilia, a completely autonomous CBF decay estimation system. We also collected a new dataset containing videos of beating ciliated cells, made available to the Scientific Community on IEEE Dataport.
The videos were acquired by an expert cytologist using a microscope (with 1000x magnification) equipped with a digital camera.
The dataset contains 119 videos of beating ciliated cells and each video exhibits a variable number of ciliated cells, varying from 1 to more than 5 (in contexts where many cells are amassed). The average video duration in the dataset is 93 seconds, with an upper bound of 352 seconds and the shortest being only 3 seconds long.
As underlined, the cardinality of ciliated cells recorded in each video can vary. From the 119 videos, a total of 246 clearly visible strias were reported, labeled, and manually extracted by the authors on each frame of the videos.
In addition to the number of cells and video duration, the videos differ from each other in terms of frame rate with an average of 23.63 Fps and and standard deviation of ± 2.54 Fps and in terms of resolution.
The video frames are also already extracted, augmented, and split in three partitions to train, validate, and test eventual object detection systems, alongside the bounding boxes for the cells manually extracted by the authors on each frame.
The dataset contains 119 videos of beating ciliated cells which can be used for research on CBF estimation. The dataset is also already partitioned into training, validation, and test sets containing 89 (185), 18 (36), and 12 (25) videos (cells) respectively; frames from this video are already extracted, and augmented and each one comes with its annotation file.
The videos were acquired by an expert cytologist using a Nikon Eclipse 600 (with 1000x magnification) microscope equipped with a camera model MD6iS (Sony IMX236 Sensor).
Each video exhibits a variable number of ciliated cells, varying from 1 to more than 5 (in contexts where many cells are amassed), while the average video duration in the dataset is 93 seconds, with an upper bound of 352 seconds and the shortest being only 3 seconds long.
In addition to the number of cells and video duration, the videos differ from each other in terms of frame rate with an average of 23.63 Fps and a standard deviation of ± 2.54 Fps, and in terms of resolution.
IMPORTANT NOTE: The annotation of the cells was carried out by experienced personnel according to their interpretation.
The acquisition and preparation of this dataset have required a lot of work without any remuneration. We provide it also free of charge, but we ask those who intend to use our dataset the courtesy to cite the following papers (thanks in advance):
- Dimauro G., Barbaro N., Camporeale M.G., Fiore V., Gelardi M., Scalera M., DeepCilia: automated, Deep Learning based engine for precise Ciliary Beat Frequency estimation, Biomedical Signal Processing and Control. https://doi.org/10.1016/i.bspc.2023.105808
- G. Dimauro, F. Girardi, M. Gelardi, V. Bevilacqua, e D. Caivano, «Rhino-Cyt: A System for Supporting the Rhinologist in the Analysis of Nasal Cytology», in Intelligent Computing Theories and Application, 2018, vol. 10955 LNCS, pp. 619–630. doi: 10.1007/978-3-319-95933-7_71
- G. Dimauro, F. Girardi, D. Caivano, e L. N. Colizzi, «Personal Health E-Record - Toward an enabling Ambient Assisted Living Technology for communication and information sharing between patients and care providers», in Ambient Assisted Living, 2019, vol. 544. doi: 10.1007/978-3-030-05921-7_39
FURTHER DETAILS
The dataset contains a folder called “video” with the 119 videos in mp4 format and a csv file named “cilia.csv” that specifies:
- Number of ciliated cells in the video;
- Framerate of the video;
- Number of frames in the video;
- Duration of the video (in seconds);
- Resolution of the video.
In the folder called “frames” are stored the frames of the video, already extracted and preprocessed.
After partitioning the videos into training, validation, and test sets containing 89 (185), 18 (36), and 12 (25) videos (cells) respectively, the frames from this video were converted to grayscale, resized to 640x640 resolution and then underwent a data augmentation step that applied one operation between shearing (both vertical and horizontal) and random rotations in the range of ± 180 degrees.
The “frames” folder thus contains two subfolders, one called “images” and one called “annotations”; both folders contain 3 more subfolders called “train”, “validation” and “testing” each one containing respectively 36287 – 7216 - 5340 frames/txt files.
Each frame in the “images” folder has its corresponding annotation file in the “annotations” folder, the correspondence between a frame and its annotation can be detected since the two share the same name.
An annotation txt file contains a row for each bounding box (in the YOLO format) in the frame. In each row there are 5 values separated by an empty space; the values respectively represent:
- The class of the bounding box (always 0 since only strias are annotated)
- The x and y coordinates of the center of the bounding box (normalized)
- The horizontal and vertical dimensions of the bounding box (normalized)
More from this Author
Dataset Files
- frames folder frames.zip (1.52 GB)
- videos folder videos.zip (13.32 GB)