Datasets
Standard Dataset
Cerebral Blood Flow Classification Dataset Using Ultra-Wideband RF Sensors with Statistical and Autoencoder-Based Features
- Citation Author(s):
- Submitted by:
- Usman Anwar
- Last updated:
- Wed, 02/12/2025 - 11:32
- DOI:
- 10.21227/zxey-1y31
- Data Format:
- Research Article Link:
- License:
 196 Views
196 Views- Categories:
- Keywords:
Abstract
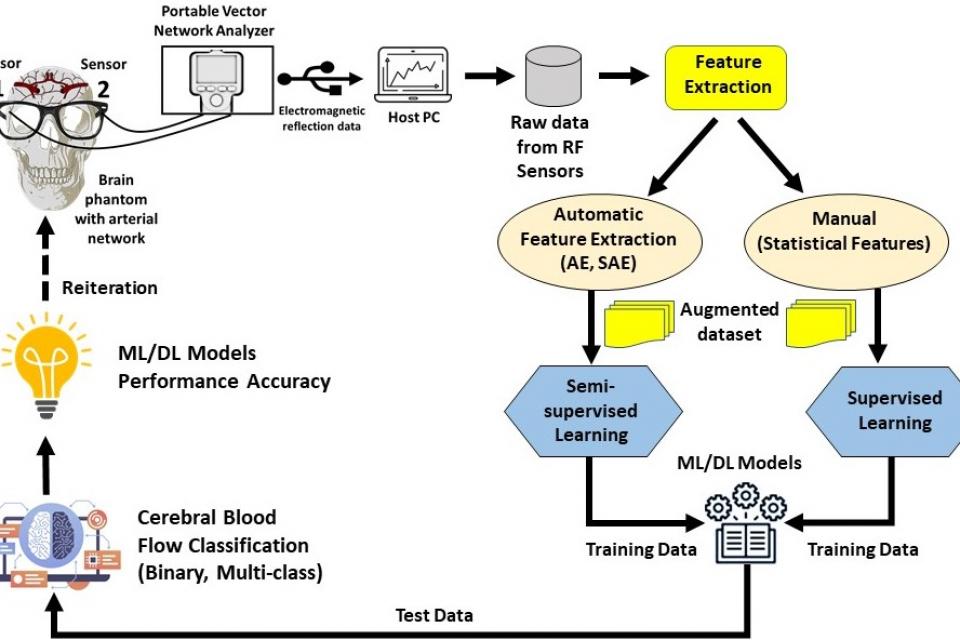
This dataset provides measurements of cerebral blood flow using Radio Frequency (RF) sensors operating in the Ultra-Wideband (UWB) frequency range, enabling non-invasive monitoring of cerebral hemodynamics. It includes blood flow feature data from two arterial networks, Arterial Network A and Arterial Network B. Statistical features were manually extracted from the RF sensor data, while autonomous feature extraction was performed using a Stacked Autoencoder (SAE) with architectures such as 32-16-32, 64-32-16-32-64, and 128-64-32-16-32-64-128. To expand the feature dataset, Gaussian noise-based feature augmentation with a mean of 0.2 and variance of 0 was applied to both statistical and autonomously extracted features. The dataset supports binary and multi-class classification. In the binary scheme, 10ml and 90ml blood flow values are labeled as abnormal, while 50ml is normal. For multi-class classification, measurements are categorized separately for each arterial network, where 10ml-A, 90ml-A, and 50ml-A represent Arterial Network A, and 10ml-B, 90ml-B, and 50ml-B correspond to Arterial Network B.
Experimental Setup:
Cerebral blood flow data was collected using Radio Frequency (RF) sensors operating in the Ultra-Wideband (UWB) frequency range to enable non-invasive monitoring of cerebral hemodynamics. The experimental setup consisted of an RF transmitter (Tx) and receiver (Rx) positioned at a fixed distance to optimize signal transmission and minimize interference. These sensors captured variations in blood flow by detecting subtle physiological changes as RF signals penetrated the skull and biological tissues.
Feature Extraction and Augmentation:
The recorded RF signals were stored in .CSV format and processed for feature extraction. Statistical features were manually extracted to capture variations in cerebral blood flow, while autonomous feature extraction was performed using a Stacked Autoencoder (SAE) with a structure of 32-16-32. To enhance the feature dataset size, Gaussian noise-based augmentation with a mean of 0.2 and variance of 0 was applied to both statistical and autonomously extracted features. This augmentation expanded the feature space, improving model generalization.
Classification and Data Structure:
The dataset supports both binary and multi-class classification.
- Binary Classification: Blood flow values of 10 ml and 90 ml are labeled as abnormal, while 50 ml is labeled as normal.
- Multi-Class Classification: The dataset consists of six classes, distinguishing between Arterial Network A and Arterial Network B:
- Arterial Network A: 10ml-A, 90ml-A, 50ml-A
- Arterial Network B: 10ml-B, 90ml-B, 50ml-B
The dataset is organized into two main folders:
- Statistical Feature Data:
- Contains two .xlsx files:
- data_0.2GaussianNoise-BinaryClass.xlsx
- data_0.2GaussianNoise-Multiclass.xlsx
- The statistical features include mean, standard deviation, quartile deviation, range, skewness, and kurtosis.
- Contains two .xlsx files:
- Stacked Autoencoder (SAE) Data:
- Contains three .xlsx files:
- Small or Arterial Network A – Binary Class: Represents binary autonomous feature data extracted using the SAE (32-16-32) and expanded via Gaussian noise augmentation.
- Big or Arterial Network B – Binary Class: Contains feature data acquired using the same methodology.
- SAE Multiclass Feature Data Gaussian Augmented: Represents six-class autonomous feature data expanded through Gaussian noise augmentation.
- Contains three .xlsx files:
Use:
This dataset serves as a foundation for developing machine learning models for cerebral blood flow classification, offering potential applications in non-invasive cerebrovascular health monitoring and the early detection of neurological disorders.
Dataset Files
- Cerebral Blood Flow Classification Dataset.zip (337.47 kB)






Comments
Kindly allow us using the dataset for educational purpose. We will acknoledge you in our publication/project.