UMMC ER-IHC Breast Histopathology Whole Slide Image and Allred Score

- Citation Author(s):
-
Wan Siti Halimatul Munirah Wan Ahmad
(Faculty of Engineering, Multimedia University, 63100 Cyberjaya, Malaysia)
Mohammad Faizal Ahmad Fauzi(Faculty of Engineering, Multimedia University, 63100 Cyberjaya, Malaysia)
Jenny Tung Hiong Lee (Department of Pathology, Sarawak General Hospital, 93586 Kuching, Sarawak, Malaysia)See Yee Khor (Hospital Seberang Jaya, Jalan Tun Hussein Onn, 13700 Seberang Jaya, Penang, Malaysia)Lai Meng Looi(Department of Pathology, University Malaya Medical Center, 59100 Kuala Lumpur, Malaysia)
Fazly Salleh Abas(Faculty of Engineering and Technology, Multimedia University, 75450 Ayer Keroh, Melaka, Malaysia)
Afzan Adam(Center for Artificial Intelligence Technology, Faculty of Information Science and Technology, Universiti Kebangsaan Malaysia, 43600 Bangi, Malaysia)
- Submitted by:
- Wan Siti Halimatul Munirah Wan Ahmad
- Last updated:
- DOI:
- 10.21227/9gbq-gz50
- Research Article Link:
 1249 views
1249 views
- Categories:
- Keywords:
Abstract
This dataset contains 37 estrogen receptor immunohistochemistry (ER-IHC) whole slide images (WSIs) obtained from Universiti Malaya Medical Centre (UMMC), Malaysia. The WSI is scanned using 3DHistech Pannoramic DESK at 20x magnification with an approximate dimension of 80,000 pixels width and 200,000 pixels height per WSI. These 37 WSIs have Allred scoring by the collaborating pathologists with a breakdown of 17 ER-negative (4 score of 0; 13 score of 2), and 20 ER-positive (12 score of 3; 5 score of 7; 3 score of 8) with regions annotated by the pathologist to assist computational method in obtaining Allred scores. Image name with related score is listed in Table 5 of our paper, and the WSI annotations (referred as ROI-WSI) are included in this dataset using WKT format (POLYGON ((x1 y1, x2 y2, x3 y3, ..., xn yn, x1 y1))).
Instructions:
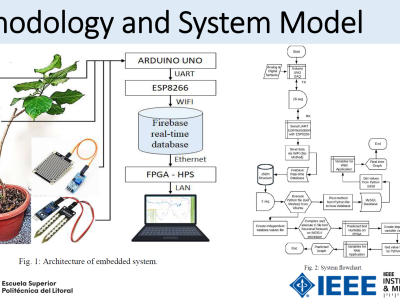
This dataset contains 37 ER-IHC whole slide images in MIRAX (.mrxs) format, at 20x magnification.
The ground truth include:
1. CSV file (ROI-WSI_annotations - Public.csv) containing cancerous tissue regions (ROI-WSI) for each WSI, annotated by our pathologists, to determine the Allred score for the particular case. The CSV file is structured as below. In the example below, 3 of the ROI-WSIs belong to the same image (4301099).
"ID" "Image" "Area" "Perimeter" "WKT"
"2762934" "4301099" "220521.7832" "1.948586768" "POLYGON ((40017.59995117188 97956.80001220704 40088.00009765625 97956.80001220704 40107.20004882813 97963.2000366211 .......))"
"2762753" "4301099" "1032907.126" "5.452907696" "POLYGON ((39766.4 102127.9996826172 39779.20004882813 102140.79973144532 39830.4 102166.4000732422 .......))"
"2762497" "4301099" "1217459.108" "6.690521135" "POLYGON ((43696.000292968754 104688.00002441407 43734.400195312504 104688.00002441407 43734.400195312504 104662.39992675782 .......))"
"ID" is the ROI-WSI annotation ID
"Image" is the WSI name (as published in the paper)
"Area" is the ROI-WSI area in micron², µ²
"Perimeter" is ROI-WSI perimeter in mm
"WKT" is the coordinates of ROI-WSI polygon (https://en.wikipedia.org/wiki/Well-known_text_representation_of_geometry)
2. The Allred score for each of the 37 WSIs are listed in Table 5 of our paper.
The segmented nuclei count shown in Table 1 is extracted using Stardist-HE pretrained model. The count for each WSI is bounded within the ROI-WSIs of the particular WSI.
If you use this dataset in any way, please cite and ensure ethical attribution of the dataset to our paper using the following citation:
W.S.H.M.W. Ahmad, M.F.A. Fauzi, M.J. Hasan, Z.U. Rehman, J.T.H. Lee, S.Y. Khor, L.M. Looi, F.S. Abas, A. Adam, E.W.L. Chan, and S. Kamata, Multi-configuration analysis of DenseNet architecture for whole slide image scoring of ER-IHC, IEEE Access, 2023.
Dataset Files
- Thumbnail images for all 37 WSIs with delineated ROI-WSI annotations in each WSI. (Size: 45.59 MB)
- CSV file containing coordinates for ROI-WSI (cancerous tissue regions) for each WSI to assist in computational method. (Size: 6.46 MB)
- Zip file containing all 37 WSIs in MIRAX format (.mrxs) and their respective folders. (Size: 16.59 GB)