Combining machine learning and metabolomics to identify weight gain biomarkers
- Citation Author(s):
-
Flavia L Dias-Audibert (UNICAMP)Diogo N Oliveira (UNICAMP)Jeany Delafiori (UNICAMP)Carlos F O R Melo (UNICAMP)Tatiane M Guerreiro (UNICAMP)Mohamed Z Dabaja (UNICAMP)Flavia T Rosa (UniFil)Diego L Petenuci (UEL)Maria A E Watanabe (UEL)Licio A Velloso (UNICAMP)Anderson R Rocha (UNICAMP)Rodrigo R Catharino (UNICAMP)
- Submitted by:
- Luiz Claudio Navarro
- Last updated:
- DOI:
- 10.21227/k446-fp12
- Data Format:
 1545 views
1545 views
- Categories:
- Keywords:
Abstract
This dataset was used in the article "Dias-Audibert FL, Navarro LC, de Oliveira DN, Delafiori J, Melo CFOR, Guerreiro TM, Rosa FT, Petenuci DL, Watanabe MAE, Velloso LA, Rocha AR and Catharino RR (2020) Combining Machine Learning and Metabolomics to Identify Weight Gain Biomarkers. Front. Bioeng. Biotechnol. 8:6. doi: 10.3389/fbioe.2020.00006", open access available at: https://doi.org/10.3389/fbioe.2020.00006.
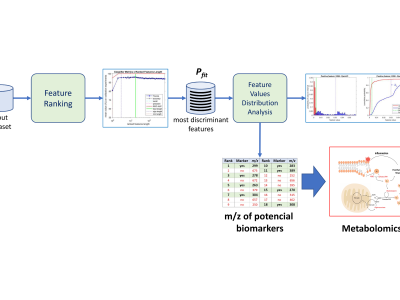
Weight gain is a metabolic disorder that often culminates in the development of obesity and other comorbidities such as diabetes. Obesity is characterized by the development of a chronic, subclinical systemic inflammation, and is regarded as a remarkably important factor that contributes to the development of such comorbidities. Therefore, laboratory methods that allow the identification of subjects at higher risk for severe weight-associated morbidity are of utter importance, considering the health and safety of populations. This contribution analyzed the plasma of 180 Brazilian individuals, equally divided into a eutrophic control group and case group, to assess the presence of biomarkers related to weight gain, aiming at characterizing the phenotype of this population. Samples were analyzed by mass spectrometry and most discriminant features were determined by a machine learning approach using Random Forest algorithm. Five biomarkers related to the pathogenesis and chronicity of inflammation in weight gain were identified. Two metabolites of arachidonic acid were upregulated in the case group, indicating the presence of inflammation, as well as two other molecules related to dysfunctions in the cycle of nitric oxide (NO) and increase in superoxide production. Finally, a fifth case group marker observed in this study may indicate the trigger for diabetes in overweight and obesity individuals. The use of mass spectrometry combined withmachine learning analyses to prospect and characterize biomarkers associated with weight gain will pave the way for elucidating potential therapeutic and prognostic targets.
Instructions:
WGMSML-Data folder contains the mass spectra input data for the Matlab scripts which are in WGMSML-MATLAB-SourceCode folder. WGMSML-ExecutionLogsAndPlots contains logs and plots generated by the execution of the Matlab code over the input data. Main scripts are enumerated in the order of execution.







