DIC images
- Citation Author(s):
- Submitted by:
- Sabyasachi Mukhopadhyay
- Last updated:
- DOI:
- 10.21227/dzsk-3d85
- Data Format:
 48 views
48 views
- Categories:
- Keywords:
Abstract
Early-stage cervical cancer is characterized by morphological changes in cells, which are effectively identified through biopsy techniques with high diagnostic accuracy. However, biopsies can be expensive and occasionally painful, with diagnosis reports often taking days or weeks to generate. These delays and costs create significant barriers for underprivileged women with limited access to timely and affordable healthcare. Our Cervi- ImagingDiag framework and app provide a painless, cost-efficient, and accessible solution, delivering diagnostic reports within seconds. The app’s lightweight design ensures compatibility with limited-resource devices while safeguarding sensitive patient data and maintaining privacy. Our proposed framework and app integrate the lightweight federated CerviImagingYOLO architecture for cervical cancer prediction in the early stages, along with the CerviImagingLangChain framework to provide detailed diagnostic explanations. The lightweight federated CerviImagingYOLO model achieved an 81% accuracy rate, outperforming lightweight federated YOLO versions YOLOv5 (80%), YOLOv6 (76.47%), YOLOv8 (75.20%), YOLOv9 (80.00%), YOLOv10 (75.70%) and other lightweight federated architectures like MobileNetV3 (61.48%), SqueezeNet (71.66%), and EfficientNet (56.30%). The CerviImagingLangChain framework enhances patient understanding through detailed explanations. The app allows doctors to upload differential interference contrast images, review automated diagnoses, and share treatment plans, while patients receive diagnosis reports, schedule appointments, and access consultations.
Instructions:
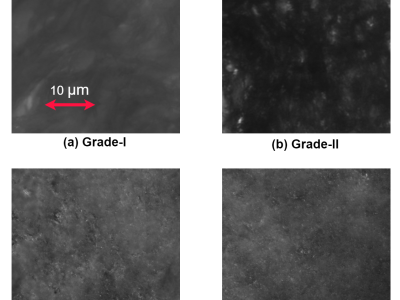
The clinical trial took place at the Super Speciality Hospital, GSVM Medical College in Kanpur, where tissue samples were collected from both healthy participants and patients with varying grades of cervical tissue, following approval from the ethical clearance committee. The trial included 4 normal individuals, 12 patients with Grade-I cervical tissue, 4 patients with Grade-II tissue, and 6 patients with Grade-III tissue, with patient ages ranging from 35 to 60 years. The excised tissues underwent a standardized histological preparation process, which involved fixation, dehydration, wax embedding, and sectioning with a rotary microtome to a thickness of approximately 5 μm, and lateral dimensions of about 4 mm × 6 mm, followed by dewaxing. Using a 60X magnification, a CCD camera was utilized to capture images through the differential interference contrast (DIC) microscopy setup. The images were captured using a CCD camera with ORCA-ERG specifications from Hamamatsu, featuring a resolution of 1344 × 1024 pixels and a pixel size of 6.45 μm. So far, there are DIC images of 102 samples of healthy or normal tissue, 88 samples of Grade-I, 167 samples of Grade-II, and 119 samples of Grade-III cervical precancerous tissue from different parts of the patients’ cervix. As part of the pre-processing by Roboflow, pixel data undergoes auto-orientation (with EXIF orientation removed), resizing to 256×256, and auto-contrast adjustment using adaptive equalization. Roboflow offers various data augmentation techniques, such as horizontal and vertical flipping, rotations (clockwise, counter-clockwise, upside-down), random cropping, rotation at varied angles, horizontal and vertical shear, and random adjustments to brightness and exposure. These augmentations were applied to the database, enhancing its suitability for modern deep learning applications. Consequently, the centralized DIC image database now includes 66,128 samples: 14,040 samples of normal tissues, 12,336 samples of Grade-I, 23,116 samples of Grade-II, and 16,636 samples of Grade-III cervical precancerous tissues. To strengthen privacy protection for medical data, different nodes or devices within the same local network handle data distribution. Each node contains approximately 7,360 datasets, including 1,560 normal samples, 1,376 Grade-I samples, 2,572 Grade-II samples, and 1,852 Grade-III samples.







simulate date