Dermatology Image and Text Dataset for AI-Powered Diagnosis and RAG-Based Medical Support
- Citation Author(s):
- Submitted by:
- Emre Olca
- Last updated:
- DOI:
- 10.21227/0ex1-ag48
 37 views
37 views
- Categories:
- Keywords:
Abstract
This dataset has been compiled and derived from publicly available dermatological image collections, including the ISIC 2018 Skin Lesion Dataset and the Atlas Dermatology archive. It comprises 49,100 high-resolution, anonymized images categorized into 32 classes, including 31 dermatological diseases and an additional “Unknown” class to improve real-world generalization. Each image is labeled based on expert classification standards and curated for deep learning applications.
In addition to visual data, the dataset integrates a text corpus composed of medical literature related to each disease class. These documents have been segmented into smaller text chunks and transformed into semantic vector representations using OpenAI embeddings. This dual structure enables both image-based disease classification and Retrieval-Augmented Generation (RAG)-based contextual medical support, allowing for reproducible research in multimodal AI-driven diagnostics.
This dataset is intended for non-commercial academic use and follows appropriate ethical guidelines. It supports research in medical computer vision, explainable AI, and hybrid decision support systems.
Instructions:
Title: Dermatology Image and Text Dataset for AI-Powered Diagnosis and RAG-Based Support
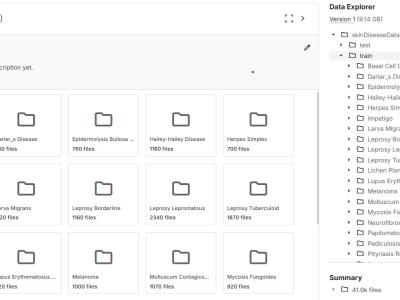
1. Dataset Contents
The dataset consists of two main components:
- Image Data
- Format: .jpg, .png
- Total Images: 49,100
- Number of Classes: 32 (31 skin diseases + 1 “Unknown”)
- Example classes: Melanoma, Psoriasis, Vitiligo, Basal Cell Carcinoma, etc.
- Source: ISIC 2018 Dataset, Atlas Dermatology
- Images are labeled and balanced across classes when possible.
- Text Data (Medical Literature)
- Format: .pdf (original), .txt (processed)
- Each file corresponds to a specific disease class
- Documents are split into semantic chunks (e.g., 512 characters)
- Each chunk is vectorized using OpenAI Embeddings and stored in .json or .csv
2. Folder Structure
Dermatology_Dataset/
│
├── images/
│ ├── Melanoma/
│ ├── Psoriasis/
│ ├── Vitiligo/
│ └── ...
│
├── documents/
│ ├── Melanoma.txt
│ ├── Psoriasis.txt
│ └── ...
│
├── vectors/
│ ├── melanoma_vectors.csv
│ ├── psoriasis_vectors.csv
│ └── ...
│
└── metadata/
└── class_labels.csv
3. Data Fields and Definitions
- image_id: Unique identifier for each image
- class_label: Disease name (e.g., Vitiligo)
- chunk_id: ID of a semantic chunk (text)
- vector_embedding: 1536-dimension float vector (OpenAI embedding output)
4. Usage Instructions
1. Image Classification Task
- Use images/ folder for training deep learning models
- Each subfolder corresponds to a class label
- Recommended input format: 224x224 RGB, normalized
2. Text Processing and RAG
- Use documents/ or vectors/ to integrate RAG pipelines
- Combine with user query embeddings for cosine similarity search
- Ideal for Retrieval-Augmented Generation or question answering systems
3. Evaluation
- Apply stratified k-fold validation due to class imbalance
- Performance metrics: Accuracy, Precision, Recall, F1-score, Confusion Matrix
5. License & Ethics
- License: Research Use Only (non-commercial)
- Ethics: All images are anonymized. No personal or identifiable data is included.