Dataset for Advanced Plaque Modeling for Atherosclerosis Detection using Molecular Communication
- Citation Author(s):
-
Pit Hofmann (Deutsche Telekom Chair of Communication Networks, Technische Universität Dresden, Dresden, Germany)Alexander Wietfeld (Chair of Communication Networks, Technical University of Munich, Munich, Germany)Jonas Fuchtmann (MITI, Klinikum rechts der Isar, Technical University Munich, Munich, Germany)Pengjie Zhou (Deutsche Telekom Chair of Communication Networks, Technische Universität Dresden, Dresden, Germany)Ruifeng Zheng (Deutsche Telekom Chair of Communication Networks, Technische Universität Dresden, Dresden, Germany)Juan Cabrera (Deutsche Telekom Chair of Communication Networks, Technische Universität Dresden, Dresden, Germany)Frank H.P. Fitzek (Deutsche Telekom Chair of Communication Networks, Technische Universität Dresden, Dresden, Germany; Centre for Tactile Internet with Human-in-the-Loop (CeTI), Dresden, Germany)Wolfgang Kellerer (Chair of Communication Networks, Technical University of Munich, Munich, Germany)
- Submitted by:
- Pit Hofmann
- Last updated:
- DOI:
- 10.21227/mn8d-0h55
 83 views
83 views
- Categories:
- Keywords:
Abstract
This dataset contains the simulation results for the microfluidic molecular simulation for advanced plaque modeling using the OpenFOAM MPPIC solver. In addition to the simulation results, first simulation results for fluid-solid interaction are uploaded using the solids4foam toolbox.
Publication:
Alexander Wietfeld, Pit Hofmann, Jonas Fuchtmann, Pengjie Zhou, Ruifeng Zheng, Juan A. Cabrera, Frank H.P. Fitzek, and Wolfgang Kellerer, “Advanced Plaque Modeling for Atherosclerosis Detection Using Molecular Communication,” submitted to International Communications Conference 2025, SAC — Molecular, Biological, and Multi-Scale Communications, June 2025.
Brief Explanation of the OpenFOAM Structure:
- File for visualization and post-processing of simulation results in, e.g., Paraview: foam.foam
- Mesh created with, e.g., Salome: 4.unv (For a description of the numbering, please see below.)
- A system directory is designated for configuring the parameters related to the solution procedure. This directory includes at least three essential files:
- controlDict: This file is responsible for establishing run control parameters, encompassing aspects such as the start and end time, time step settings, and parameters for data output.
- fvSchemes: Within this file, users can dynamically select discretization schemes employed during the solution process.
- fvSolution: In this file, users can define equation solvers, tolerance levels, and various algorithm controls required for the current run.
- Within the constant directory, comprehensive information about the case mesh is stored. This directory also hosts subdirectories like polyMesh, which house detailed descriptions of the mesh, along with files that define the physical properties relevant to the specific application. An example of such a file is transportProperties.
- The time directories house individual data files pertaining to specific fields. This data can encompass two distinct types: either the initial values and boundary conditions, which necessitate user specification to define the problem, or results generated by OpenFOAM and saved to files. The initial conditions are typically stored in a directory named "0".
Instructions:
Package:
- Simulation results for the channel length of 50 millimeters, using the Casson fluid model*: d50_casson_OpenFOAM.zip
- Template for an easy start with the OpenFOAM MPPIC solver (including a .txt file with instructions to run the case) and Python file for post-processing: see https://ieee-dataport.org/documents/dataset-simulation-microfluidic-molecular-communication-using-openfoam
- Simulation results for the channel length of 50 millimeters, without a plaque formation, for the fluid-solid interaction using the solids4foam toolbox: d50_FSI_solids4foam.zip. Further information can be found here: https://www.solids4foam.com/
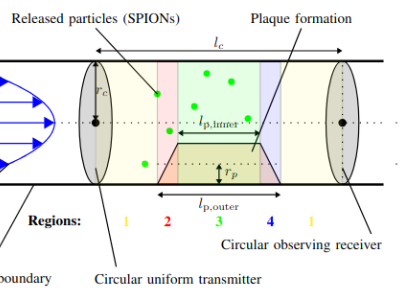
* Naming of the various simulation scenarios: Given the example "Plaque_laminar_1_varying_016", "Plaque" denotes the existing plaque in the model, "laminar" assumes laminar flow, "1" denotes the plaque size (1 -> no plaque, 4 -> r_p = 0.25 x r_c, 7 -> r_p = 0.5 x r_c, 10 -> r_p = 0.75 x r_c), "varying" denotes the given pulsatile inlet velocity boundary condition, and "016" the release time of the particles (016 -> t_release = 0.16 s, 04 -> t_release = 0.4 s, 09 -> t_release = 0.9 s).
For further instructions, please refer to the publication.