Cell line gene expression data

- Citation Author(s):
-
Jingyang Ge
- Submitted by:
- Gejingyang Ge
- Last updated:
- DOI:
- 10.21227/z97t-9856
 149 views
149 views
- Categories:
- Keywords:
Abstract
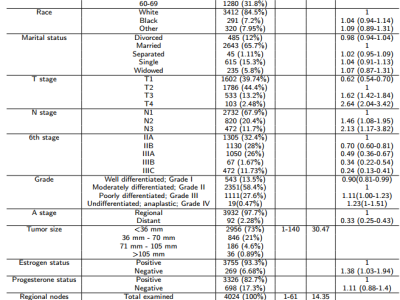
The table contains the standard symbols for genes, with each symbol corresponding to a specific gene. The GENE_title column provides the full names of the genes. The subsequent numerical values represent experimental data related to the genes, with each column indicating the gene expression levels (e.g., FPKM values from RNA-Seq data) under different experimental conditions, samples, or states. The magnitude of these values reflects variations in gene expression across the corresponding samples or conditions. This data is typically used for gene function studies, differential expression analysis, or sample classification. It can also serve as the initial embedding features for cancer cell lines in the task of predicting anticancer drug synergy.
Instructions:
The table contains the standard symbols for genes, with each symbol corresponding to a specific gene. The GENE_title column provides the full names of the genes. The subsequent numerical values represent experimental data related to the genes, with each column indicating the gene expression levels (e.g., FPKM values from RNA-Seq data) under different experimental conditions, samples, or states. The magnitude of these values reflects variations in gene expression across the corresponding samples or conditions. This data is typically used for gene function studies, differential expression analysis, or sample classification. It can also serve as the initial embedding features for cancer cell lines in the task of predicting anticancer drug synergy.