Other
Research data for Mitigating Safety Risks in Information Systems: A Self-Adaptive Approach Research Data. A controlled experiment was conducted to determine whether SAS, aware of interruptions, improves effectiveness, efficiency, situational awareness, and usability compared to non-adaptive systems in information systems. A total of 30 participants contributed to the controlled experiment.
- Categories:
 34 Views
34 Views
This dataset comprises user-generated content from Stack Overflow, including post bodies, post tags, and user engagement metrics such as upvotes and downvotes. The data was collected from the stack exchange explorer based on user defined categories and other criteria like reputation and badges as explained in our work. It was collected to support research in technology and emotion analysis, focusing on understanding user interactions and sentiments within online communities.
- Categories:
 19 Views
19 Views
Integrating multiple (sub-)systems is essential to create advanced Information Systems. Difficulties mainly arise when integrating dynamic environments, e.g., the integration at design time of not yet existing services. This has been traditionally addressed using a registry that provides the API documentation of the endpoints.
- Categories:
 19 Views
19 Views
Silicon nitride has greatly improved the versatility of integrated photonics technologies due to its wide transparency window, compatibility with existing CMOS-foundry infrastruc ture, and its potential for extremely low propagation loss. However, its low refractive index reduces mode confinement, increasing the minimum device footprint of components such as the widely-adopted ring resonator filter, resulting in a height ened need for reduced-bending architecture to maintain high integration density.
- Categories:
 10 Views
10 Views
Silicon nitride has greatly improved the versatility of integrated photonics technologies due to its wide transparency window, compatibility with existing CMOS-foundry infrastruc ture, and its potential for extremely low propagation loss. However, its low refractive index reduces mode confinement, increasing the minimum device footprint of components such as the widely-adopted ring resonator filter, resulting in a height ened need for reduced-bending architecture to maintain high integration density.
- Categories:
 11 Views
11 Views
This paper presents a Tube-based Robust Model Predictive Control (Tube-RMPC) strategy for autonomous vehicle control, designed to address model parameter uncertainties and variations in road-tire adhesion coefficients under complex driving conditions. The proposed approach enhances the representation of vehicle dynamics by introducing a unified vehicle-tire modeling framework, capturing nonlinear characteristics more effectively.
- Categories:
 21 Views
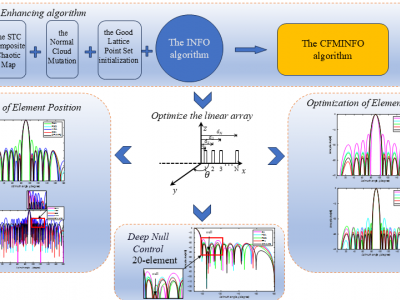
21 ViewsAbstract—This manuscript introduces the Chaos Fusion Mutation INFO algorithm (CFMINFO), which integrates multiple strategies and updates vector positions through three core processes. These processes incorporate Good Point Set initialization, Sine-Tent-Cosine (STC) chaotic parameterization, and Normal Cloud Mutation strategies. The algorithm is characterized by its simplicity, rapid convergence, and ability to avoid local optima. To validate its performance, CFMINFO is applied to the optimization of linear arrays.
- Categories:
 104 Views
104 Views
The dataset contains a revised version of the psychophysical color difference Munsell Re-renotation dataset.
Munsell Re-renotation is a psychophysical dataset describing large color differences, featuring 2986 colors characterized by standard colorimetric coordinates (x, y, Y) and coordinates within the Munsell system (H, V, C).The Munsell Re-renotation iteration enhances the uniformity of the system compared to its predecessor, the Munsell Renotation dataset.
- Categories:
 35 Views
35 Views
Transformers, especially dry-type transformers, which nowadays are going to be employed instead of oil-type transformers, are one of the major equipment in the generation, transmission and distribution network of electric energy. The transformer insulation strength may reduce due to partial discharge (PD) occurrence, and this can finally result in its insulation failure.
- Categories:
 14 Views
14 Views
- Categories:
 24 Views
24 Views