Datasets
Standard Dataset
CrossDocked2020
- Citation Author(s):
- Submitted by:
- Zhang Xiang
- Last updated:
- Mon, 11/11/2024 - 11:13
- DOI:
- 10.21227/45c9-vg74
- Data Format:
- License:
 235 Views
235 Views- Categories:
- Keywords:
Abstract
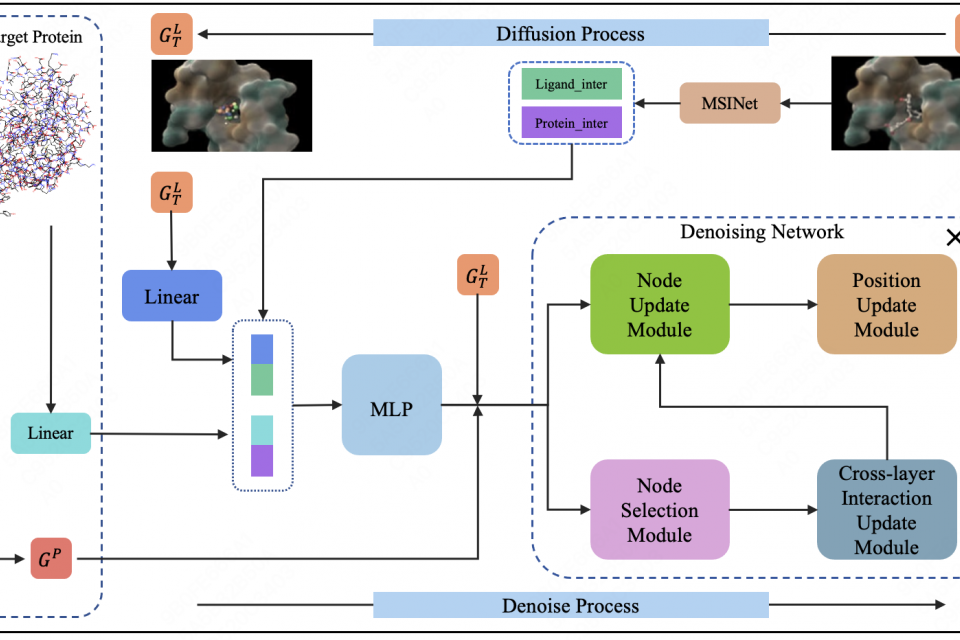
In structure-based drug design (SBDD), a major challenge is generating high-affinity 3D ligand molecules that can effectively bind to specific protein targets, which requires accurately capturing complex protein-ligand interactions. Although existing diffusion models have demonstrated potential in molecular generation tasks, they often struggle with accurately capturing the complex interactions between proteins and ligands. To address this problem, we propose MSIDiff, a multi-stage interaction-aware diffusion model for protein-specific molecular generation. MSIDiff uses the pre-trained model MSINet to extract real protein-ligand interaction information during the initial diffusion stage and incorporates this information into the reverse process to ensure that the generated molecules exhibit accurate interaction relationships with target proteins. Through a scoring mechanism, MSIDiff filters key nodes to extract crucial protein-ligand interaction data and uses the cross-layer interaction update module with GRU to recursively integrate information from different denoising stages, enabling effective cross-layer information transmission. Experimental results on the CrossDocked2020 dataset show that MSIDiff can generate molecules with more realistic 3D structures and higher binding affinity to protein targets, achieving an Avg. Vina Score of up to -6.36, while maintaining appropriate molecular properties.
In structure-based drug design (SBDD), a major challenge is generating high-affinity 3D ligand molecules that can effectively bind to specific protein targets, which requires accurately capturing complex protein-ligand interactions. Although existing diffusion models have demonstrated potential in molecular generation tasks, they often struggle with accurately capturing the complex interactions between proteins and ligands. To address this problem, we propose MSIDiff, a multi-stage interaction-aware diffusion model for protein-specific molecular generation. MSIDiff uses the pre-trained model MSINet to extract real protein-ligand interaction information during the initial diffusion stage and incorporates this information into the reverse process to ensure that the generated molecules exhibit accurate interaction relationships with target proteins. Through a scoring mechanism, MSIDiff filters key nodes to extract crucial protein-ligand interaction data and uses the cross-layer interaction update module with GRU to recursively integrate information from different denoising stages, enabling effective cross-layer information transmission. Experimental results on the CrossDocked2020 dataset show that MSIDiff can generate molecules with more realistic 3D structures and higher binding affinity to protein targets, achieving an Avg. Vina Score of up to -6.36, while maintaining appropriate molecular properties.
Documentation
| Attachment | Size |
|---|---|
| 10.16 KB |