Datasets
Standard Dataset
Dataset for the UNB-GCN model
- Citation Author(s):
- Submitted by:
- Gang Wang
- Last updated:
- Fri, 05/05/2023 - 01:27
- DOI:
- 10.21227/6v9g-dz81
- Data Format:
- License:
 107 Views
107 Views- Categories:
- Keywords:
Abstract
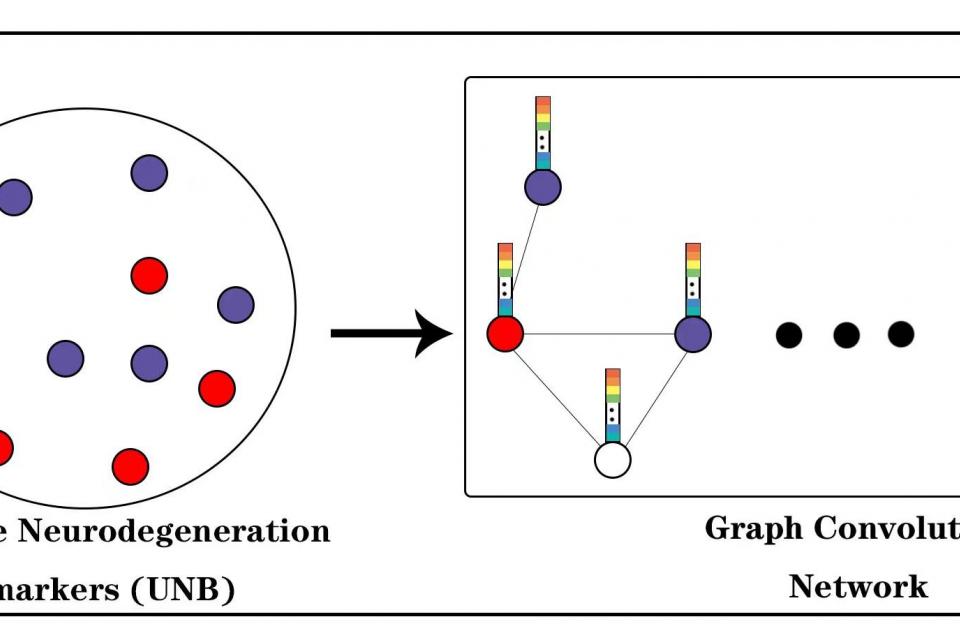
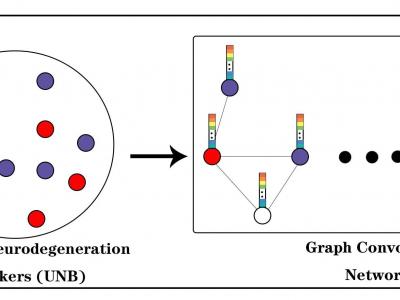
For the UNB-GCN classification model, UNB as the input feature vector to the graph structure node, phenotypic information, such as gender, age and APOE genetic information, as the weight of the edges between nodes.
Detailed instructions on how to utilize the dataset
(replace DS by the name of the dataset):
n = total number of nodes
m = total number of edges
N = number of graphs
(1) DS_A.txt (m lines)
sparse (block diagonal) adjacency matrix for all graphs,
each line corresponds to (row, col) resp. (node_id, node_id)
(2) DS_graph_indicator.txt (n lines)
column vector of graph identifiers for all nodes of all graphs,
the value in the i-th line is the graph_id of the node with node_id i
(3) DS_graph_labels.txt (N lines)
class labels for all graphs in the dataset,
the value in the i-th line is the class label of the graph with graph_id i
(4) DS_node_labels.txt (n lines)
column vector of node labels,
the value in the i-th line corresponds to the node with node_id i
There are OPTIONAL files if the respective information is available:
(5) DS_edge_labels.txt (m lines; same size as DS_A_sparse.txt)
labels for the edges in DS_A_sparse.txt
(6) DS_edge_attributes.txt (m lines; same size as DS_A.txt)
attributes for the edges in DS_A.txt
(7) DS_node_attributes.txt (n lines)
matrix of node attributes,
the comma seperated values in the i-th line is the attribute vector of the node with node_id i
(8) DS_graph_attributes.txt (N lines)
regression values for all graphs in the dataset,
the value in the i-th line is the attribute of the graph with graph_id i
More from this Author
Dataset Files
- Dataset used to classify AD vs.MCI ADMCI.rar (231.95 kB)
- Dataset used to classify AD vs.NC ADNC.rar (272.73 kB)
Documentation
| Attachment | Size |
|---|---|
| 1.41 KB |